Viral manipulation of immunity
Understanding how viruses subvert innate and adaptive antiviral responses
The Carlin Lab is located at the UCSD School of Medicine, in La Jolla, CA. Dr. Carlin's lab investigates transcriptional mechanisms that determine the outcome of host-pathogen interactions. A combination of genome-wide, molecular and genetic approaches are used to understand the molecular mechanisms by which pathogenic viruses trigger pro- and anti-microbial transcriptional programs and how they contribute to immunity or pathogenesis. This detailed understanding will provide insight into why specific pathogens cause human disease, spur development of rapid infectious disease diagnostics and help identify the optimal targets for new antimicrobials.

Understanding how viruses subvert innate and adaptive antiviral responses

We are very interested in how viruses influence regulatory mechanisms to subvert cellular defenses. This includes effects that are gene-specific, transcription factor specific as well as global effects on transcription, splicing, and RNA stability. We aim to determine if specific interactions occur universally or in a cell-type specific manner and how they contribute to virulence.
Publications/Links
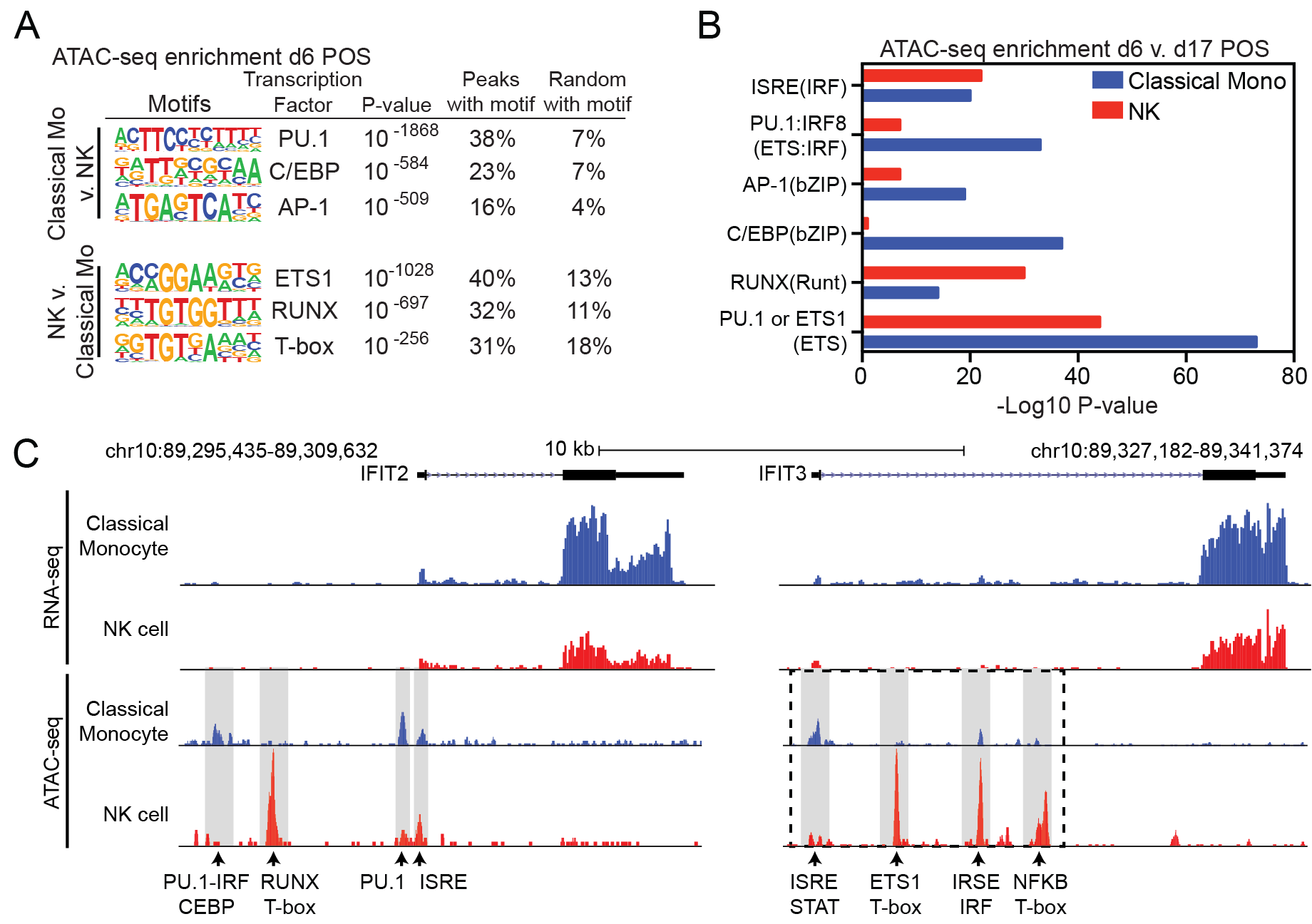
Leveraging genome-wide technologies and bioinformatics analysis to study antiviral transcriptional networks

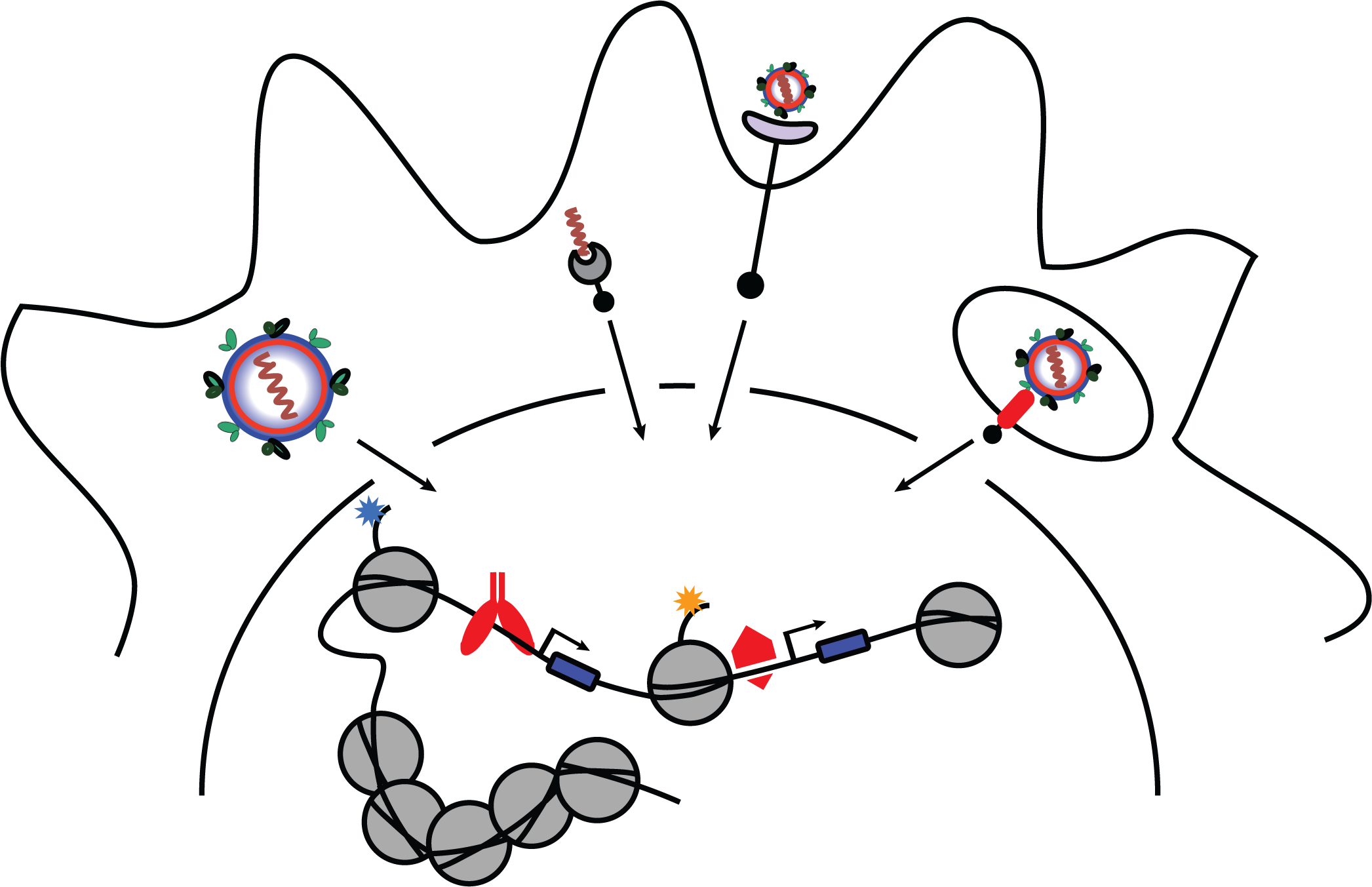
We study innate immune responses, leveraging genome-wide technologies and bioinformatics analysis to better understand transcriptional networks regulated by interferon regulatory factors (IRFs) and anti-viral pathways. We study how the interplay between key inflammatory transcription factors, such as IRFs, NF-kB, and STATs, and cell-type specific master regulators (e.g. PU.1, C/EBP) induce specific target genes and antiviral transcriptional programs.
Publications
Identifying key self and non-self recognition receptors that influence pathogen recognition, host-response and pathogen eradication.

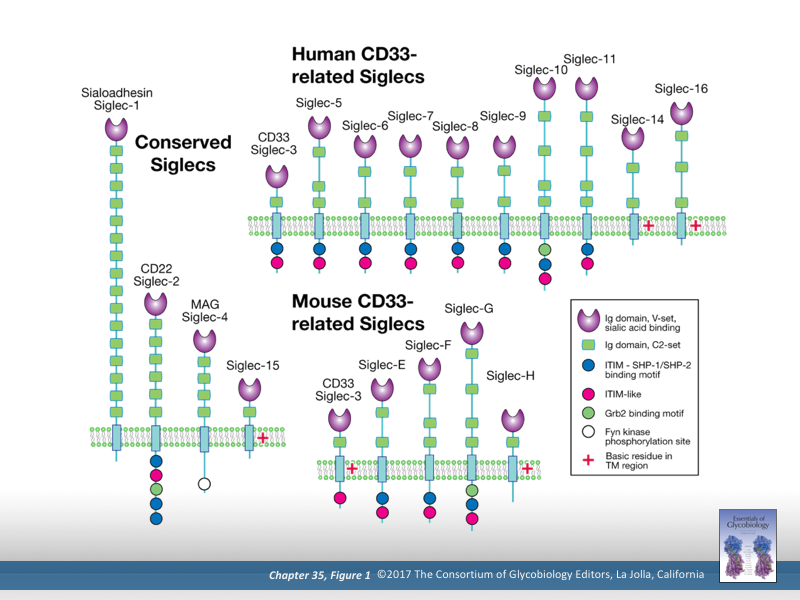
Sialic acid binding immunoglobulin type lectins (Siglecs) are expressed on immune cells and are thought to play a key role in self/non-self recognition by sensing self-associated glycans. Pathogens have evolved to exploit Siglecs in glycan-dependent and -independent manners. At the same time, CD33related Siglecs are also rapidly evolving leading to differential Siglec expression patterns within the human population. We are interested in understanding how pathogen interactions with Siglecs modulate disease pathogenesis as well as how human genetic diversity of Siglecs alters susceptibility to microorganisms. Image used with permission. Copyright Consortium of Glycobiology Editors
Publications
Developing and improving systems based approaches to analyze and integrate quantitative NGS data to understand human-viral interactions

Complementary, genome-wide methods, including CRISPR, transcriptomics and epigenomics, provide an unbiased and comprehensive way of expanding our understanding of host–viral interactions. However, many of these methods can be adapted to maximally interrogate human responses to viral infections in vivo or ex vivo. We are developing experimental and computational tools that improve sample processing and data integration to provide biologically meaningful conclusions. We work closely with the Heinz, Benner and Glass Laboratories at UCSD to develop optimized or innovative NGS protocols to more specifically interrogate host-viral interactions in humans.
Publications
Developing and optimizing antimicobials to treat emerging infectious diseases

We collaborate with chemists, microbiologists and infectious disease physicians to develop and optimize new therapeutics for emerging infectious diseases.
Publications
Leibel SL, McVicar RN, Murad R, Kwong EM, Clark AE, Alvarado A, Grimmig BA, Nuryyev R, Young RE, Lee JC, Peng W, Zhu YP, Griffis E, Nowell CJ, James B, Alarcon S, Malhotra A, Gearing LJ, Hertzog PJ, Galapate CM, Galenkamp KMO, Commisso C, Smith DM, Sun X, Carlin AF, Sidman RL, Croker BA, Snyder EY. A therapy for suppressing canonical and noncanonical SARS-CoV-2 viral entry and an intrinsic intrapulmonary inflammatory response. Proc Natl Acad Sci U S A 2024 Jul 23
Duttke SH, Guzman C, Chang M, Delos Santos NP, McDonald BR, Xie J, Carlin AF, Heinz S, Benner C. Position-dependent function of human sequence-specific transcription factors. Nature 2024 Jul 17
Ray P, Ledgerwood-Lee M, Brickner H, Clark AE, Garretson A, Graham R, Van Zant W, Carlin AF, Aronoff-Spencer ES. Design and Development of an Antigen Test for SARS-CoV-2 Nucleocapsid Protein to Validate the Viral Quality Assurance Panels. Viruses 2024 April 24.
Tomazini BM, Tramujas L, Medrado FA Junior, Gomes SPDC, Negrelli KL, Murinize GS, Santos RHN, Vianna BMP, Piotto BF, Veiga TS, Santos BRD, Peneluppi Horak AC, Lemos OMC, Lopes MA, Olicheski BB, Campones DL, Peixoto LAA, Basilio ADAC, Gebara OCE, Lopes ATA, Saconato H, Valeis N, Miranda TA, Laranjeira LN, Santucci EV, Carlin AF, Esko JD, Gordts PLSM, Tsimikas S, Cavalcanti AB. Halofuginone for non-hospitalized adult patients with COVID-19 a multicenter, randomized placebo-controlled phase 2 trial. The HALOS trial. PLOS ONE 2024 Feb 23
McMillan RE, Lo MK, Zhang XQ, Beadle JR, Valiaeva N, Garretson AF, Clark AE, Freshman JE, Murphy J, Montgomery JM, Spiropoulou CF, Schooley RT, Hostetler KY, Carlin AF. Enhanced broad spectrum in vitro antiviral efficacy of 3-F-4-MeO-Bn, 3-CN, and 4-CN derivatives of lipid remdesivir nucleoside monophosphate prodrugs. Antiviral Res 2023 Sep 25
Clark AE, Furst A, Sejane K, Stellwagen L, Proost M, Pride D, Smith DM, Carlin AF, Bode L. Validating Tools to Detect and Inactivate Monkeypox Virus in Human Milk. Breastfeed Med 2023 Sep 23
Carlin AF, Clark AE, Garretson AF, Bray W, Porrachia M, Santos AT, Rana TM, Chaillon A, Smith DM. Neutralizing Antibody Responses After Severe Acute Respiratory Syndrome Coronavirus 2 BA.2 and BA.2.12.1 Infection Do Not Neutralize BA.4 and BA.5 and Can Be Blunted by Nirmatrelvir/Ritonavir Treatment. Open Forum Infect Dis 2023 Mar 21
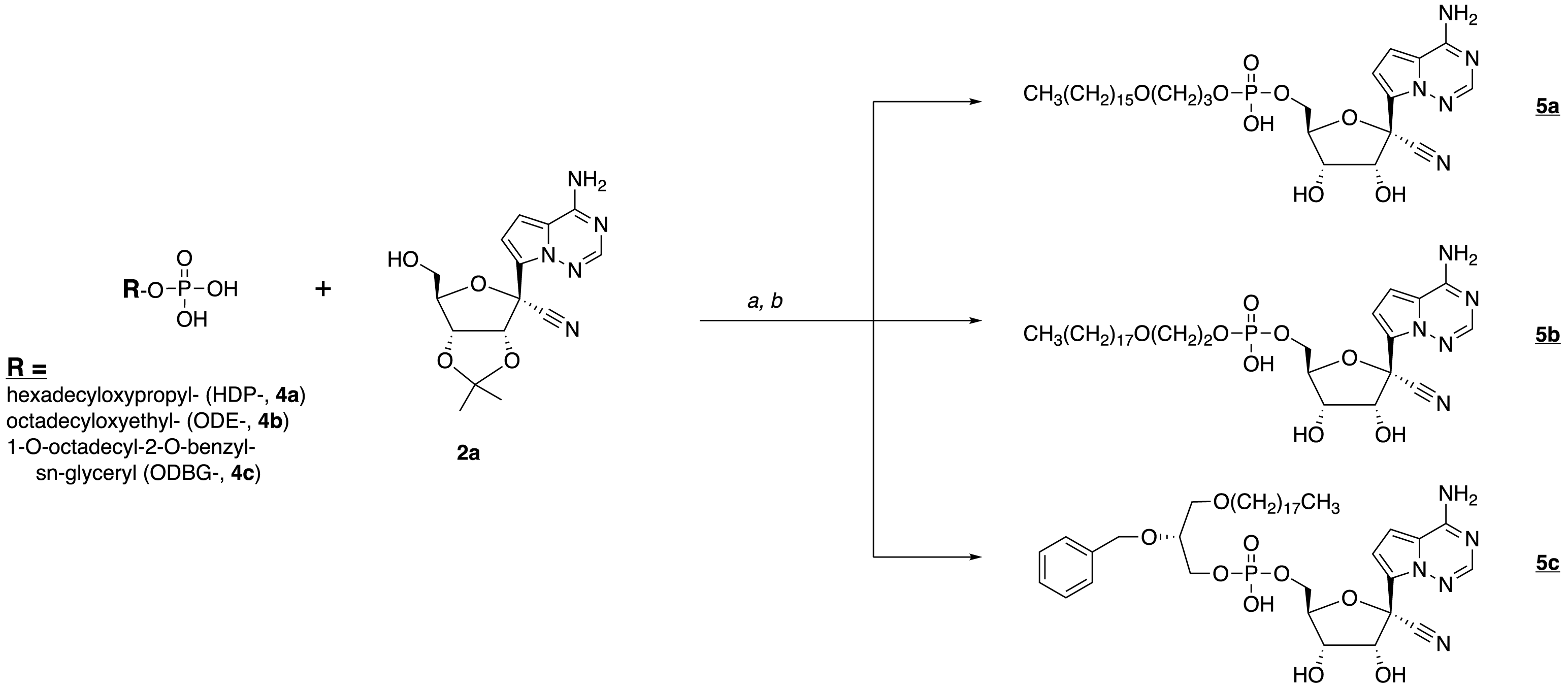
Carlin AF, Beadle JR, Clark AE, Gully KL, Moreira FR, Baric RS, Graham RL, Valiaeva N, Leibel SL, Bray W, McMillan RE, Freshman JE, Garretson AF, McVicar RN, Rana T, Zhang XQ, Murphy JA, Schooley RT, Hostetler KY. 1-O-Octadecyl-2-O-benzyl-sn-glyceryl-3-phospho-GS-441524 (V2043). Evaluation of Oral V2043 in a Mouse Model of SARS-CoV-2 Infection and Synthesis and Antiviral Evaluation of Additional Phospholipid Esters with Enhanced Anti-SARS-CoV-2 Activity. J Med Chem 2023 Apr 11
McMillan RE, Wang E, Carlin AF, Coufal NG. Human microglial models to study host-virus interactions. Exp Neurol 2023 Mar 14
Sørensen DM, Büll C, Madsen TD, Lira-Navarrete E, Clausen TM, Clark AE, Garretson AF, Karlsson R, Pijnenborg JFA, Yin X, Miller RL, Chanda SK, Boltje TJ, Schjoldager KT, Vakhrushev SY, Halim A, Esko JD, Carlin AF, Hurtado-Guerrero R, Weigert R, Clausen H, Narimatsu Y. Identification of global inhibitors of cellular glycosylation. Nat Commun 2023 Feb 20
Cheng Y, Clark AE, Yim W, Borum RM, Chang YC, Jin Z, He T, Carlin AF, Jokerst JV. Protease-Responsive Potential-Tunable AIEgens for Cell Selective Imaging of TMPRSS2 and Accurate Inhibitor Screening. Anal Chem 2023 Feb 21
Wang L, Wang S, Wu L, Li W, Bray W, Clark AE, Gonzalez GM, Wang Y, Carlin AF, Rana TM. PCIF1-mediated deposition of 5'-cap N6,2'-O-dimethyladenosine in ACE2 and TMPRSS2 mRNA regulates susceptibility to SARS-CoV-2 infection. Proc Natl Acad Sci U S A 2023 Jan 31
Branche E, Wang YT, Viramontes KM, Cuevas JMV, Xie J, Duttke SH, Garretson AF, Crames J, Spann NJ, Benner C, Shresta S, Carlin AF. Zika Virus activates SREBP2-dependent Lipid Gene Transcription to Infect Human Dendritic Cells. Nat Commun 2022 Sep 12
Alamanza G, Kouznetsova V, Clark AE, Olmedillas E, Castro A, Tsigelny IF, Wu Y, Gao GF, Burton DR, Saphire EO, Carlin AF and Zanetti M. Structure-selected RBM immunogens prime polyclonal memory responses with potent neutralizing activity. PLOS Pathogens 2022 Jul 21
Carlin AF, Clark AE, Chaillon A, Garretson AF, Bray W, Porrachi M, Santos AT, Rana TM, Smith DM. Virologic and Immunologic Characterization of COVID-19 Recrudescence after Nirmatrelvir/Ritonavir Treatment. Clin Infect Dis 2022 June 20
Duttke S, Beyhan S, Singh R, Neal S, Viriyakosol S, Fierer J, Kirkland TN, Stajich JE, Benner C, Carlin AF. Decoding Transcription Regulatory Mechanisms Associated with Coccidioides immitis Phase Transition Using Total RNA mSystems 2022 Feb 22
Schooley RT and Carlin AF, Beadle JR, Valiaeva N, Zhang XQ, Clark AE, McMillan RE, Leibel SL, McVicar RN, Xie J, Garretson AF, Smith VI, Murphy J, Hostetler KY. Rethinking Remdesivir: Synthesis, Antiviral Activity and Pharmacokinetics of Oral Lipid Prodrugs. Antimicrob Agents Chemother.2021 Jul 26
Wang L, Sievert D, Clark AE, Lee S, Federman H, Gastfriend BD, Shusta EV, Palecek SP, Carlin AF and Gleeson JG. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nature Medicine.2021 July 09
Park SH, Siddiqi H, Castro D, Angelis AD, Oom A, Stoneham C, Lewinski M, Clark AE, Croker B, Carlin AF, Guatelli J, Opella Interactions of SARS-CoV-2 envelope protein with amilorides correlate with antiviral activity. PLOS Pathogens.2021 May 18
Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, Knight R, Smith D, Wang Y, Carlin AF, Rana TM. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Reports2021 May 11
Mellott D, Tseng CT, Drelich A, Fajtova P, Chenna BC, Kostomiris D, Hsu JC, Zhu J, Taylor Z, Tat V, Katzfuss A, Li L, Giardini MA, Skinner D, Hirata K, Beck S, Carlin AF, Clark AE, Berreta L, Maneval D, Frueh F, Hurst BL, Wang H, Kocurek KI, Raushel FM, O'Donoghue A, Siqueira-Neto JL, Meek TD, McKerrow JH. A cysteine protease inhibitor blocks SARS-CoV-2 infection of human and monkey cells. ACS Chem Biol.2021 Mar 31
Singh N, Ray P, Carlin AF, Magallanes C, Morgan S, Laurent L, Aronoff-Spencer E, Hall D. Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens Bioelectron.2021 Feb 26
Tiwari SK, Wang S, Smith D, Carlin AF, and Rana T. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell derived lung and cerebral organoids. Stem Cell Reports 2021 Mar 9
Mor M, Werbner M, Alter J, Safra M, Chomsky E, Lee JC, Hada-Neeman S, Polonsky K, Nowell CJ, Clark AE, Roitburd-Berman A, Ben-Shalom N, Navon M, Rafael D, Sharim H, Kiner E, Griffis ER, Gershoni JM, Kobiler O, Leibel SL, Zimhony O, Carlin AF, Yaari G, Dessau M, Gal-Tanamy M, Hagin D, Croker BA, Freund NT. Multi-Clonal Live SARS-CoV-2 In Vitro Neutralization by Antibodies Isolated from Severe COVID-19 Convalescent Donors. PLOS Pathogens 2021 Feb 11
Clausen TM, Sandoval DR, Spliid CB, Pihl J, Painter CD, Thacker BE, Glass CA, Narayanan A, Majowicz SA, Zhang Y, Torres JL, Golden GJ, Porell R, Garretson AF, Laubach L, Feldman J, Yin X, Pu Y, Hauser B, Caradonna TM, Kellman BP, Martino C, Gordts PLSM, Leibel SL, Chanda SK, Schmidt AG, Godula K, Jose J, Corbett KD, Ward AB, Carlin AF, Esko JD. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020 Nov 12
Wang S, Li W, Hui H, Tiwari SK, Zhang Q, Croker BA, Rawlings S, Smith D, Carlin AF, Rana TM. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and coronaviruses by depleting membrane cholesterol. EMBO J. 2020 Sept 18
Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020 Jun 25
Carlin AF, Wen J, Vizcarra EA, McCauley M, Chaillon A, Akrami K, Kim C, Elong Ngono A, Lara-Marquez ML, Smith DM, Glass CK, Schooley RT, Benner C, Shresta S. A longitudinal systems immunologic investigation of acute Zika virus infection in an individual infected while traveling to Caracas, Venezuela. PLOS NTD 2018 December 31
Carlin AF and Shresta S. Genome-wide approaches to unravelling host–virus interactions in Dengue and Zika infections. Current Opinion in Virology 2018 December 18
Fowler AM, Tang WW, Young MP, Mamidi A, Viramontes KM, McCauley MD, Carlin AF, Schooley RT, Swanstrom J, Baric RS, Govero J, Diamond MS, Shresta S. Maternally Acquired Zika Antibodies Enhance Dengue Disease Severity in Mice. Cell Host and Microbe 2018 November 14
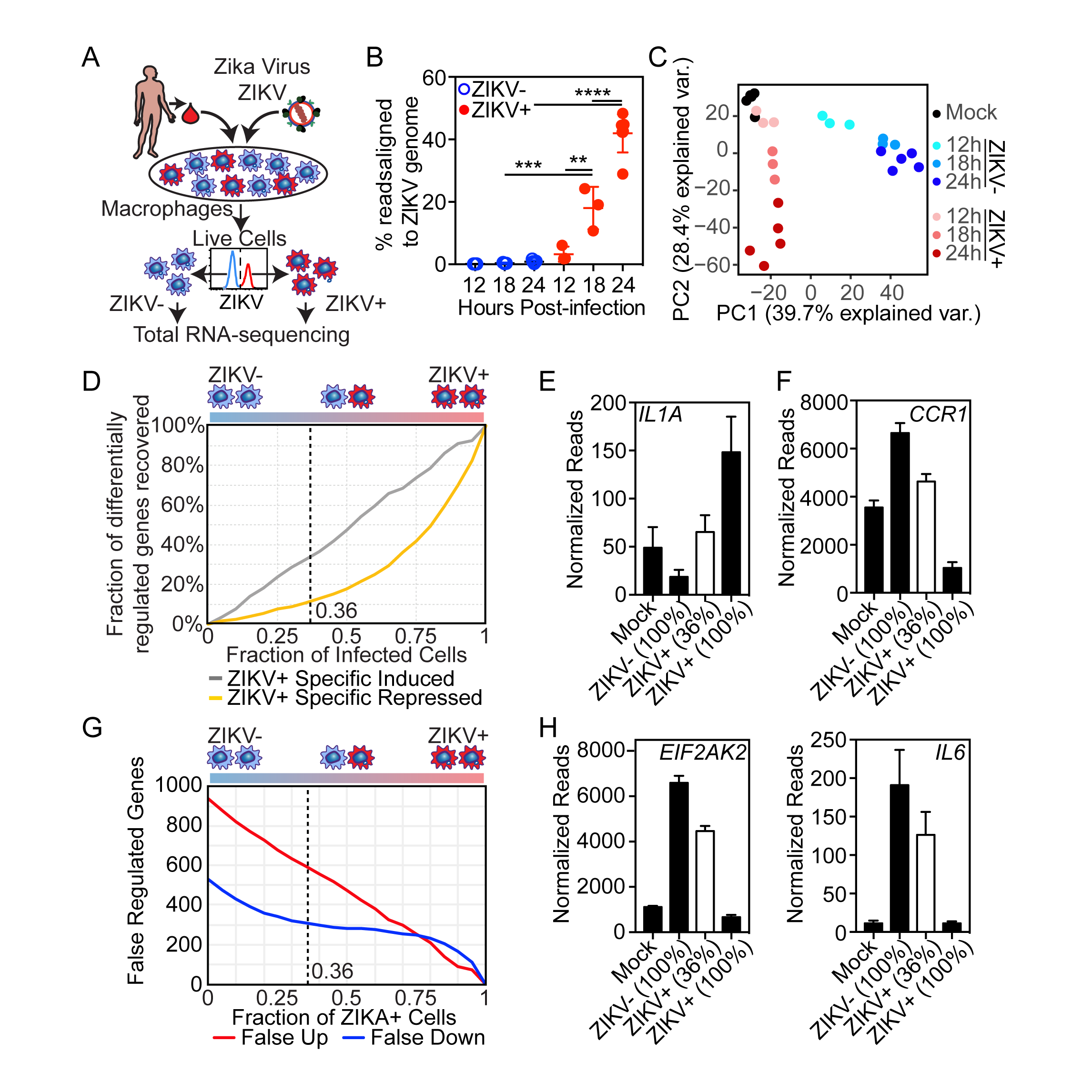
Carlin AF, Vizcarra EA, Branche E, Viramontes KM, Ley K, Heinz S, Benner C, Shresta S, Glass CK. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc Natl Acad Sci U S A. 2018 September 25
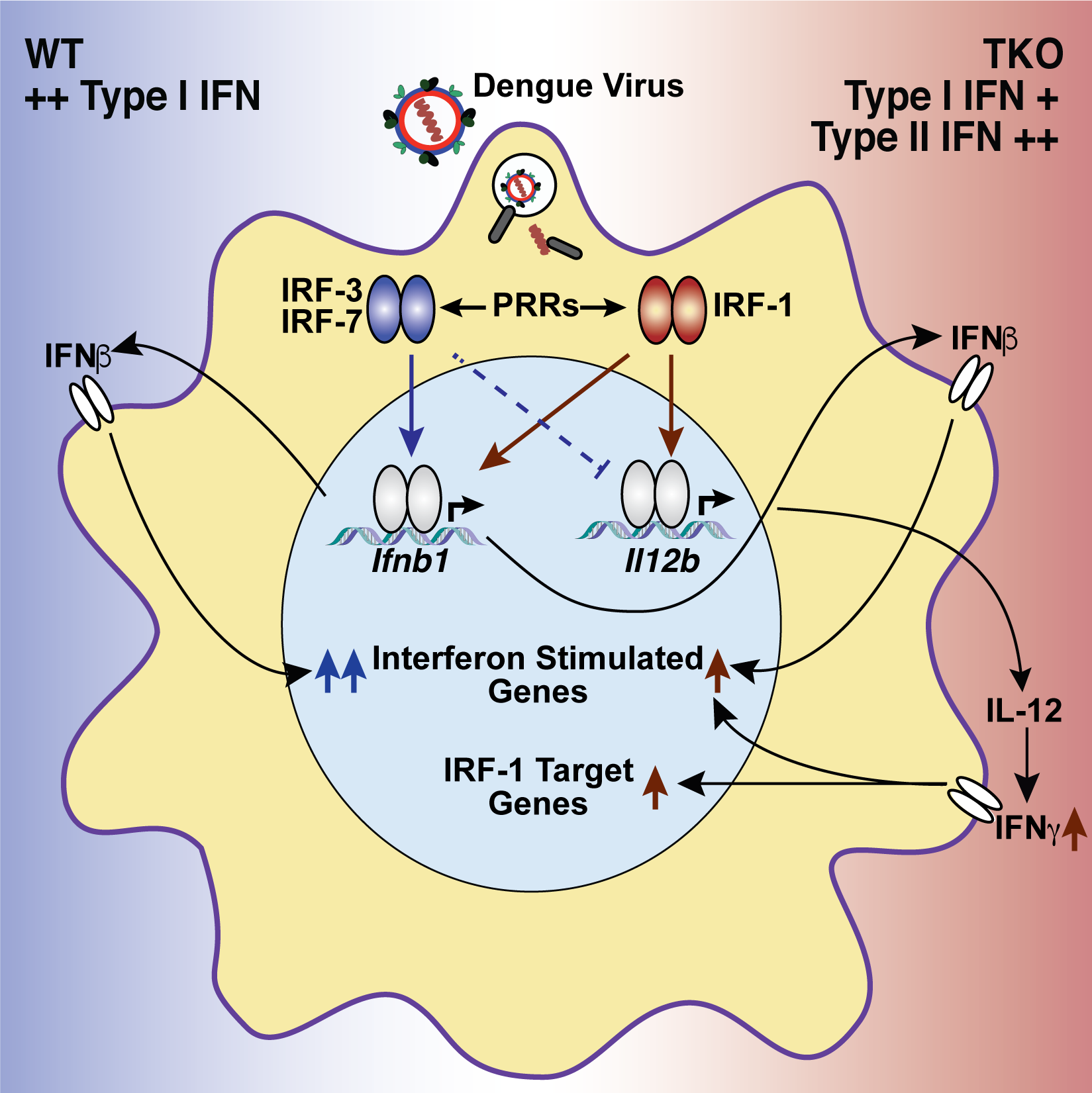
Carlin AF, Plummer EM, Vizcarra E, Sheets N, Joo Y, Tang W, Day J, Greenbaum J, Glass CK, Diamond MS, Shresta S.An IRF-3, -5, -7-independent alternative pathway of viral resistance utilizes IRF-1 to stimulate an IFN-γ response in macrophages.Cell Reports 2017 November 7
Naito-Matsui Y, Davies LRL, Takematsu H, Chou HH, Tangvoranuntakul P, Carlin AF, Verhagen A, Heyser CJ, Yoo SW, Choudhury B, Paton JC, Paton AW, Varki NM, Schnaar RL,Varki A.Physiological Exploration of the Long Term Evolutionary Selection against Expression of N-Glycolylneuraminic Acid in the Brain.J Biol Chem. 2017 Feb 17
Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, Kolar MJ, Matsuzaka T, Hayakawa S, Tao J, Kaikkonen MU, Carlin AF, Lam MT, Manabe I, Shimano H, Saghatelian A, Glass CK.SREBP1 Contributes to Resolution of Pro-inflammatory TLR4 Signaling by Reprogramming Fatty Acid Metabolism.Cell Metabolism 2017 Feb 7
Carlin AF, Aristizabal P, Song Q, Wang H, Paulson MS, Stamm LM, Schooley RT, Wyles DL.Temporal dynamics of inflammatory cytokines/chemokines during sofosbuvir and ribavirin therapy for genotype 2 and 3 hepatitis C infection.Hepatology 2015 July 3
Ali SR, Fong JJ, Carlin AF, Busch TD, Linden R, Angata T, Areschoug T, Parast M, Varki N, Murray J, Nizet V, Varki A.Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus.J Exp Med 2014 June 2
Muto J, Morioka Y, Yamasaki K, Kim M, Garcia A, Carlin AF, Varki A, Gallo RL.Hyaluronan digestion controls DC migration from the skin.J Clin Invest 2014 February 3
Shibata N*, Carlin AF*, Spann NJ, Saijo K, Morello CS, McDonald JG, Romanoski CE, Maurya MR, Kaikkonen MU, Lam MT, Crotti A, Reichart D, Fox JN, Quehenberger O, Raetz CR, Sullards MC, Murphy RC, Merrill AH Jr, Brown HA, Dennis EA, Fahy E, Subramaniam S, Cavener DR, Spector DH, Russell DW, Glass CK.25-Hydoxycholesterol activates the integrated stress response to reprogram transcription and translation in macrophages.J Biol Chem 2013 December 13
Uchiyama, S*, Carlin AF*, Khosravi A, Weiman S, Banerjee A, Quach D, Hightower G, Mitchell TJ, Doran KS, Nizet V.The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion.J Exp Med 2009 August 17
Carlin AF, Chang YC, Areschoug T, Lindahl G, Hurtado-Ziola N, King CC, Varki A, Nizet V.Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5.J Exp Med 2009 Aug 3
Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A.Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response.Blood 2009 April 2
Carlin AF, Lewis AL, Varki A, Nizet V.Group B Streptococcal Capsular Sialic Acids Interact with Siglecs (Immunoglobulin-Like Lectins) on Human Leukocytes.J Bacteriol 2007 Feb
* authors contributed equally to this work

Aaron received his BS in Microbiology and Molecular Genetics and Philosophy at UCLA before enrolling in the Medical Scientist Training Program at UCSD. He joined the laboratories of Dr. Victor Nizet and Dr. Ajit Varki to pursue his Ph.D. in Biomedical Sciences. As a graduate student, Aaron demonstrated that group B Streptococcus (GBS) utilizes molecular mimickry and direct protein interactions to engage Sialic acid-binding immunoglobulin-type lectins (Siglecs) on human innate immune cells. This interaction impairs antimicrobial functions increasing GBS pathogenesis. After completing medical school, Aaron joined the Physician Scientist Training Program at UCSD where he completed a residency in Internal Medicine and a fellowship and Chief fellowship in Infectious Diseases. Aaron joined the laboratory of Chris Glass for his post-doctoral fellowship training where he developed unbiased genome-wide methods to study how viruses subvert transcription. Aaron was appointed as an Assistant Professor at UCSD in 2019.







acarlin@ucsd.edu
(858) 246-3261
9500 Gilman Drive MC 0640
La Jolla, CA 92093-0640
George Palade Laboratories (CMM west)
GPL Room 100G
School of Medicine
University of California, San Diego