Genomics Analysis, Motif Discovery, NGS Method Development
Developing software programs for the analysis, annotation, and integration of quantitative NGS data, including RNA-Seq, ChIP-Seq, ATAC-seq
The Benner Lab is located at the UCSD School of Medicine, in La Jolla, CA. We are a genomics and bioinformatics and sequencing technology laboratory focused on decoding the regulatory functions of the genome. We use next-generation sequencing (NGS) and computational analysis to understand the mechanisms responsible for transcription and epigenetic regulation in human development and disease. Our primary focus is to elucidate enhancer function in the immune system, where we use a variety of advanced NGS techniques to characterize changes in transcription, chromatin states and genome structure during the host defense response to foreign pathogens. Due to the laboratory’s expertise in developing NGS assays and analysis algorithms, we are involved in a wide array of collaborative projects spanning cancer, metabolism, development, cellular reprogramming, and evolution.

Developing software programs for the analysis, annotation, and integration of quantitative NGS data, including RNA-Seq, ChIP-Seq, ATAC-seq

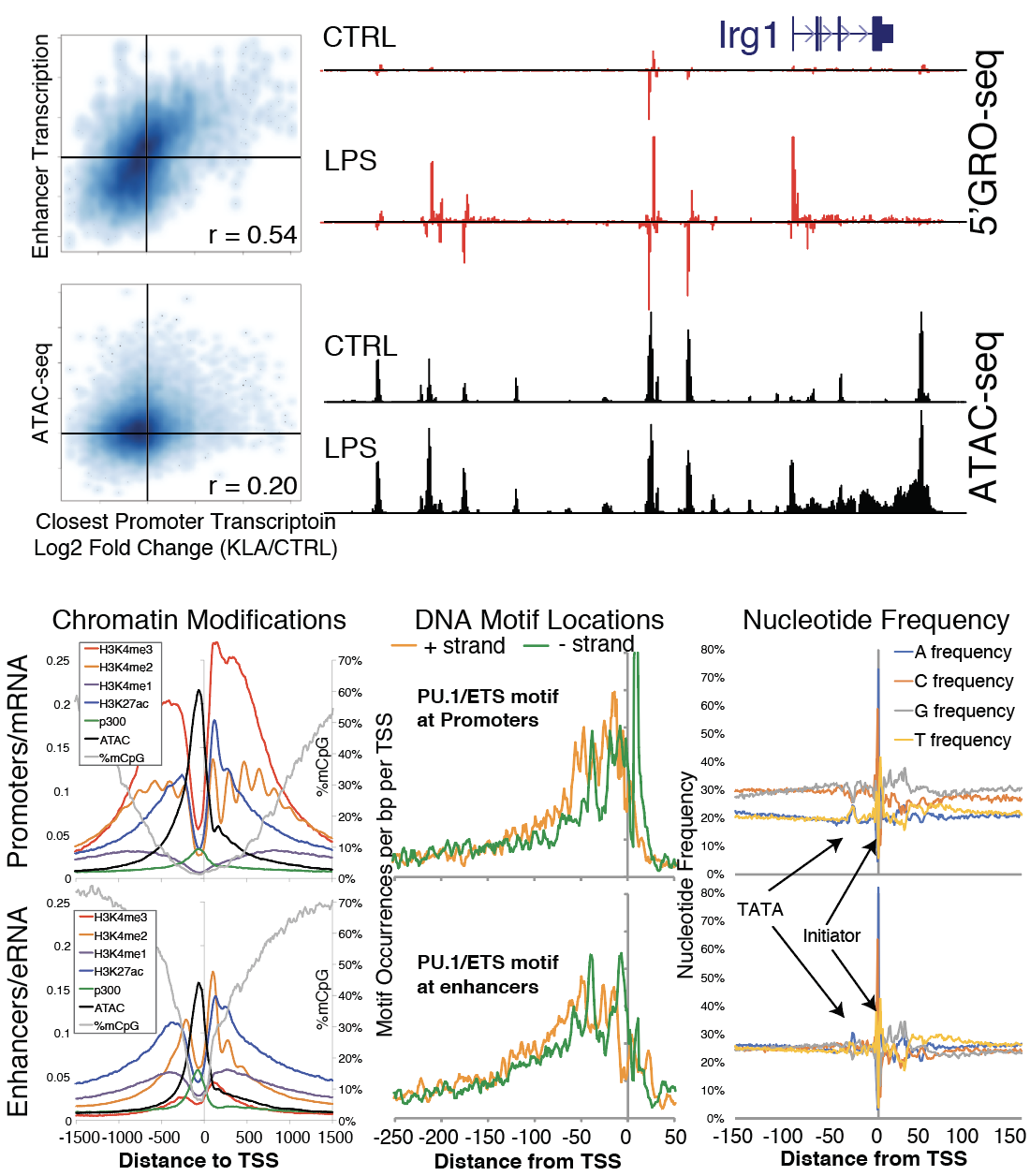
Advances in NGS technology and rapid innovation in NGS assay development have resulted in a great need for computational analysis tools capable of both processing data and integrating results toward biologically meaningful conclusions. We are focused on developing software programs for the analysis, annotation, and integration of quantitative NGS data, including RNA-Seq, ChIP-Seq, ATAC-seq. We are particularly interested in leveraging more advanced NGS assays that are capable of characterizing specific transcriptional and epigenetic states, including nascent RNA profiling (GRO-Seq) and chromatin structure (Hi-C). In order to integrate these findings with the function of non-coding DNA, we are developing tools to analyze regulatory sequences, including DNA motif discovery and the analysis of regulatory architectures. Most of our previous algorithm and software development efforts have been bundled together as part of the HOMER software suite. In addition to data analysis, we work closely with the Heinz and Glass Laboratories at UCSD to develop optimized or innovative NGS protocols to more specifically interrogate regulatory mechanisms, including the development of transcriptional initiation strategies and modified chromatin interaction capture protocols.
Publications/Links
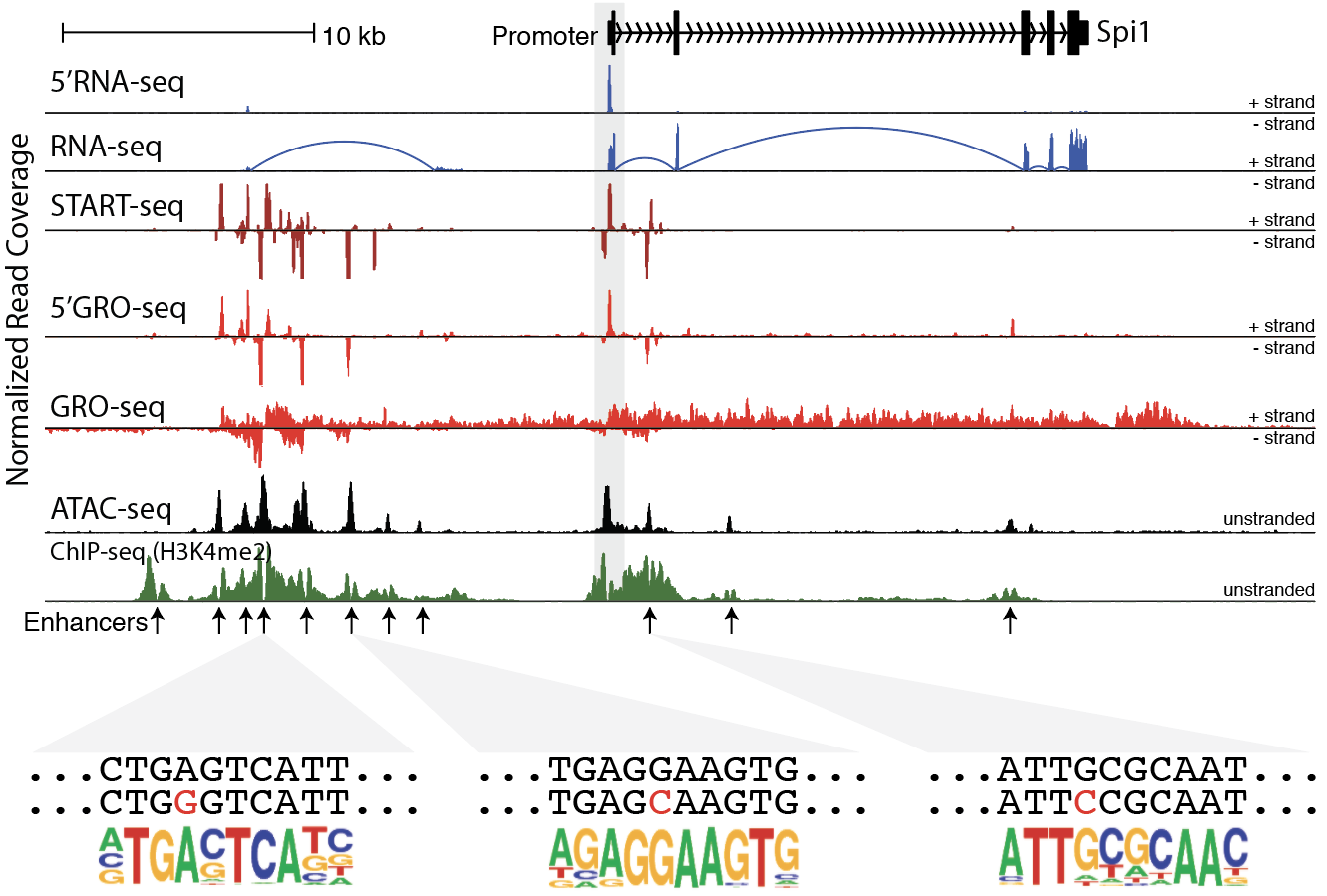
Using nascent RNA sequencing to detect transcription rates at enhancer elements to measure enhancer activity

Cell-type and stimulus specific gene expression is often modulated by promoter-distal enhancer elements, but the exact mechanisms by which these regulatory features function and influence the expression of nearby genes is poorly understood. We are interested in using nascent RNA sequencing (5’GRO-seq, GRO-Seq, START-seq) to detect unstable RNAs at enhancer elements to measure enhancer activity and study their interplay between transcription factor recruitment, epigenetic modifications, and genome structure. We study these processes within the context of cellular differentiation and innate immune signaling in the hematopoietic lineage, attempting to link the activation of enhancers with functional consequences in the immune system. We are also investigating the effect of natural and disease-associated genetic variation on enhancer function to better understand the impact that non-coding DNA has on regulatory networks.
Publications
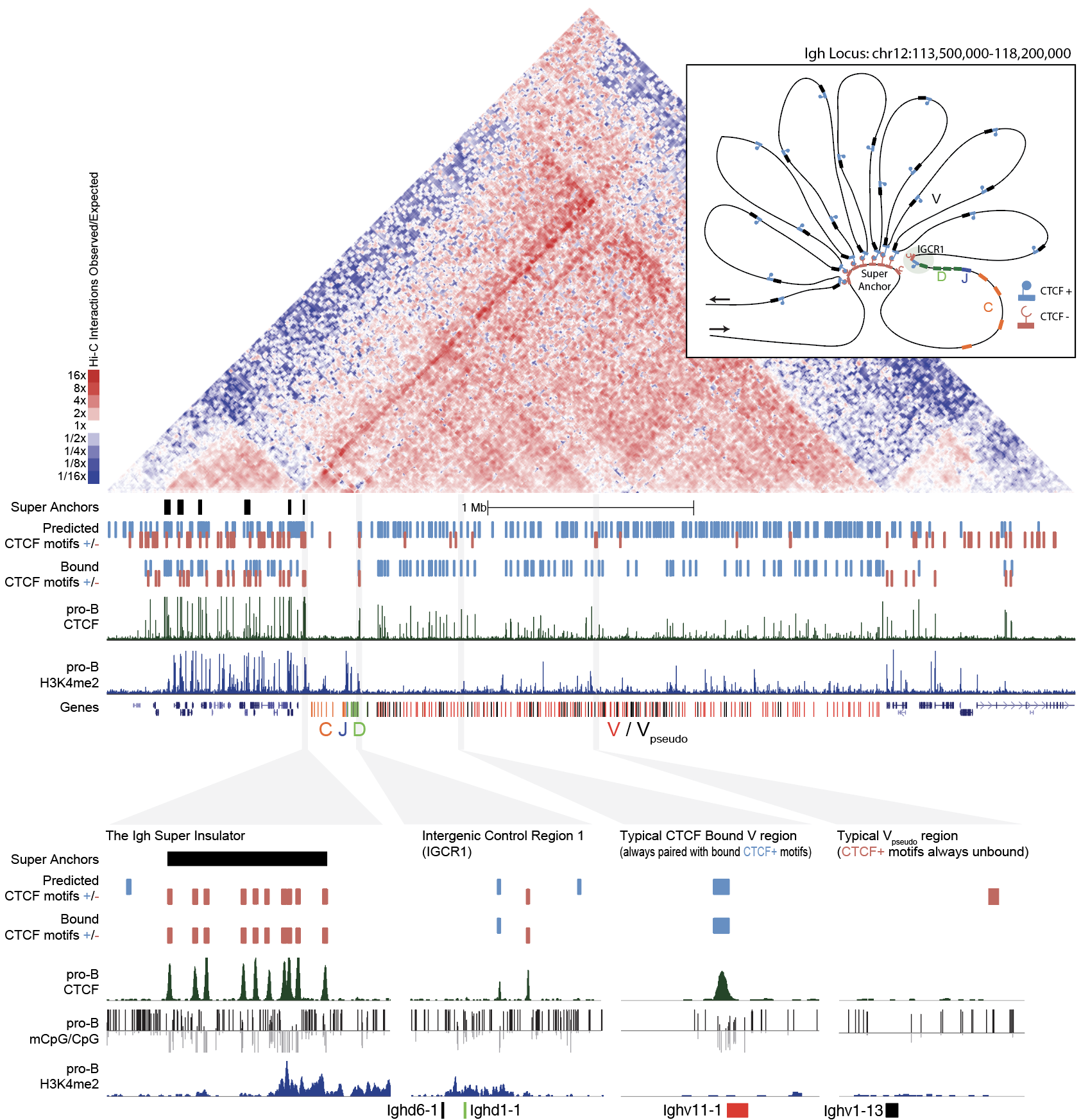
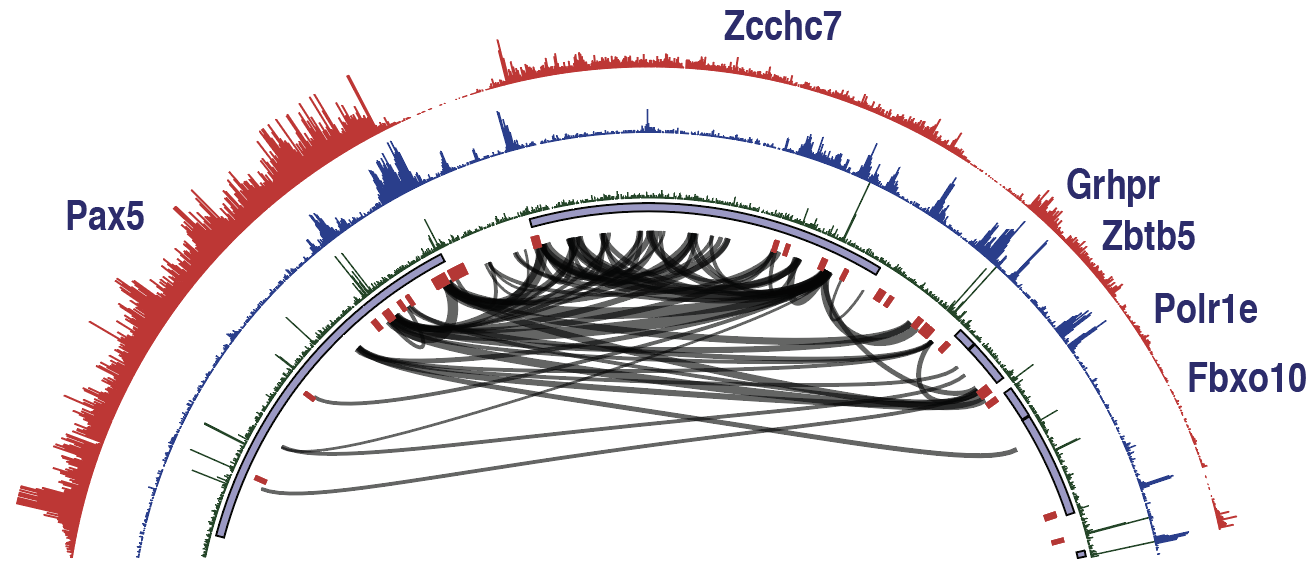
Developing software and innovative approaches to analyze Hi-C data and integrate with transcriptome and epigenetic data

The structural configuration of chromatin in the nucleus plays an important role in regulating gene expression. We have worked extensively with the Heinz laboratory at UCSD to develop software and innovative approaches to analyze Hi-C data and integrate it with transcriptome and epigenetic data. Changes in genome structure are important determinates for cellular differentiation and have important implications during the immune response. The genome structure underlying the adaptive immune system not only orchestrates tightly controlled patterns of gene expression, but must also facilitate the functional recombination of antigen receptor loci. A deep understanding of these processes is important to understand B and T cell function and how Ig and TCR loci are capable of generating diverse repertoires of receptors to combat disease.
Publications
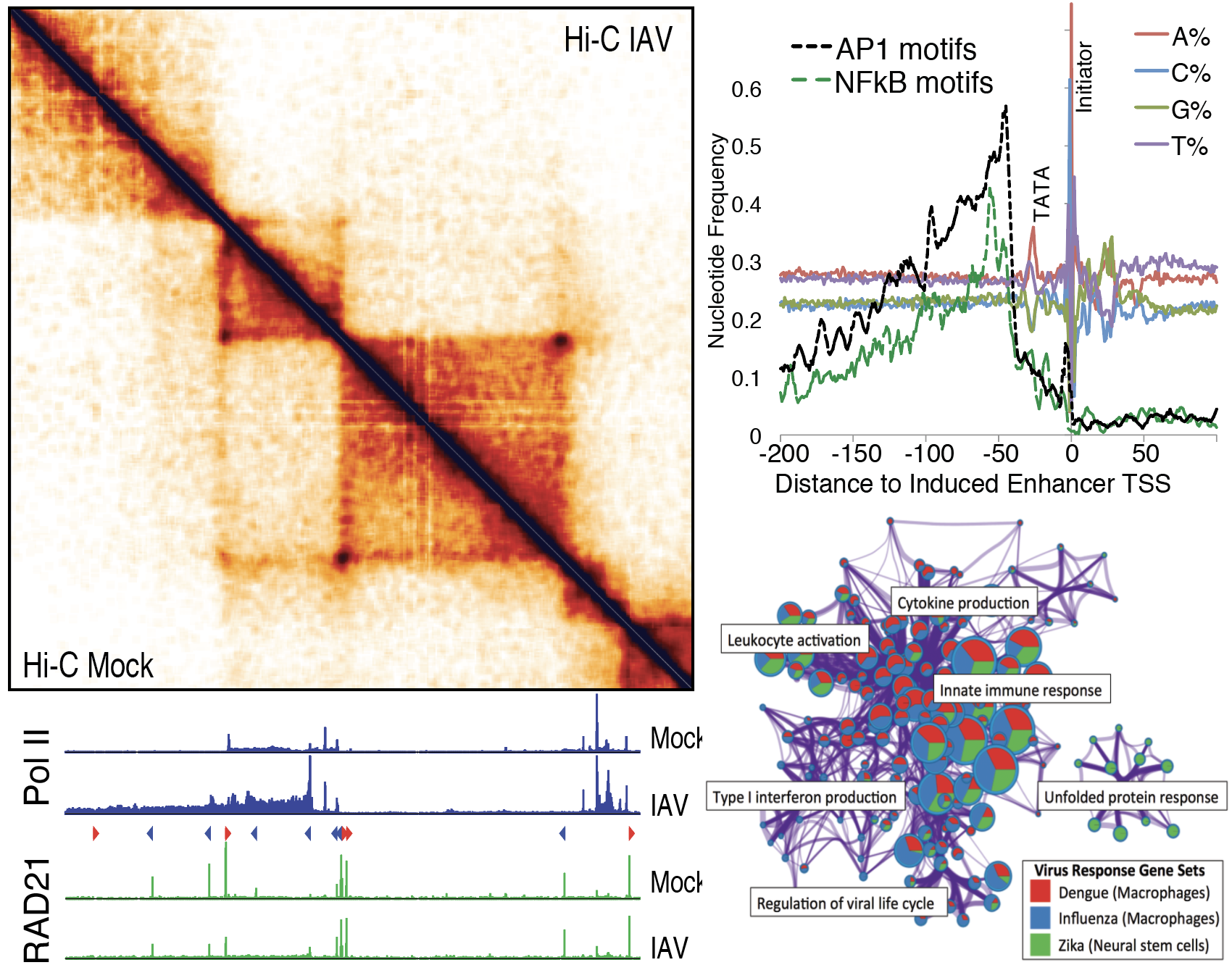
Leveraging genome-wide technologies and bioinformatics analysis to study transcriptional networks

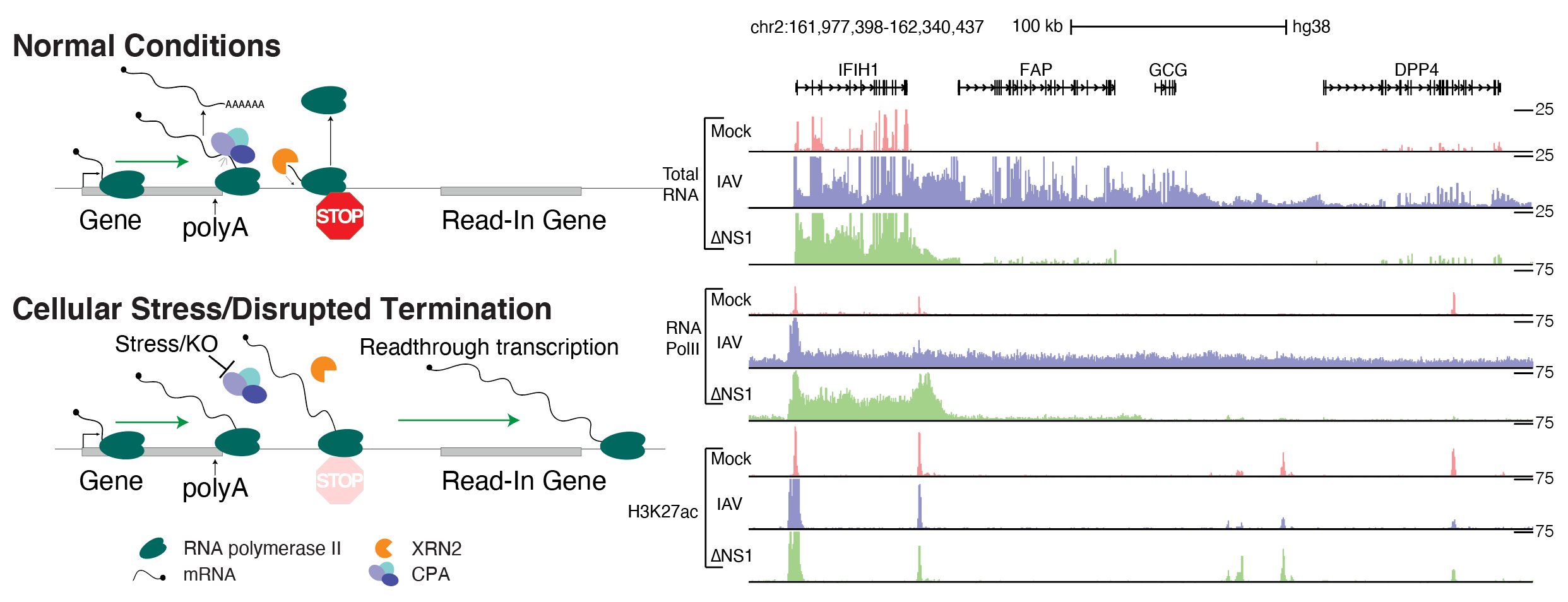
We participate in multiple projects studying innate immunity, leveraging genome-wide technologies and bioinformatics analysis to better understand transcriptional networks regulated by Toll-like receptors and anti-viral pathways. We study how the interplay between key inflammatory transcription factors (e.g., NF-kB, IRFs, STATs) and cell-type specific master regulators (e.g. PU.1, C/EBP, EBF1) induce specific target genes, and how genome structure, epigenetics, and enhancer RNAs are impacted. In particular, we are very interested in how viruses influence regulatory mechanisms to subvert cellular defenses, including global effects on transcription termination, splicing, and RNA stability.
Publications
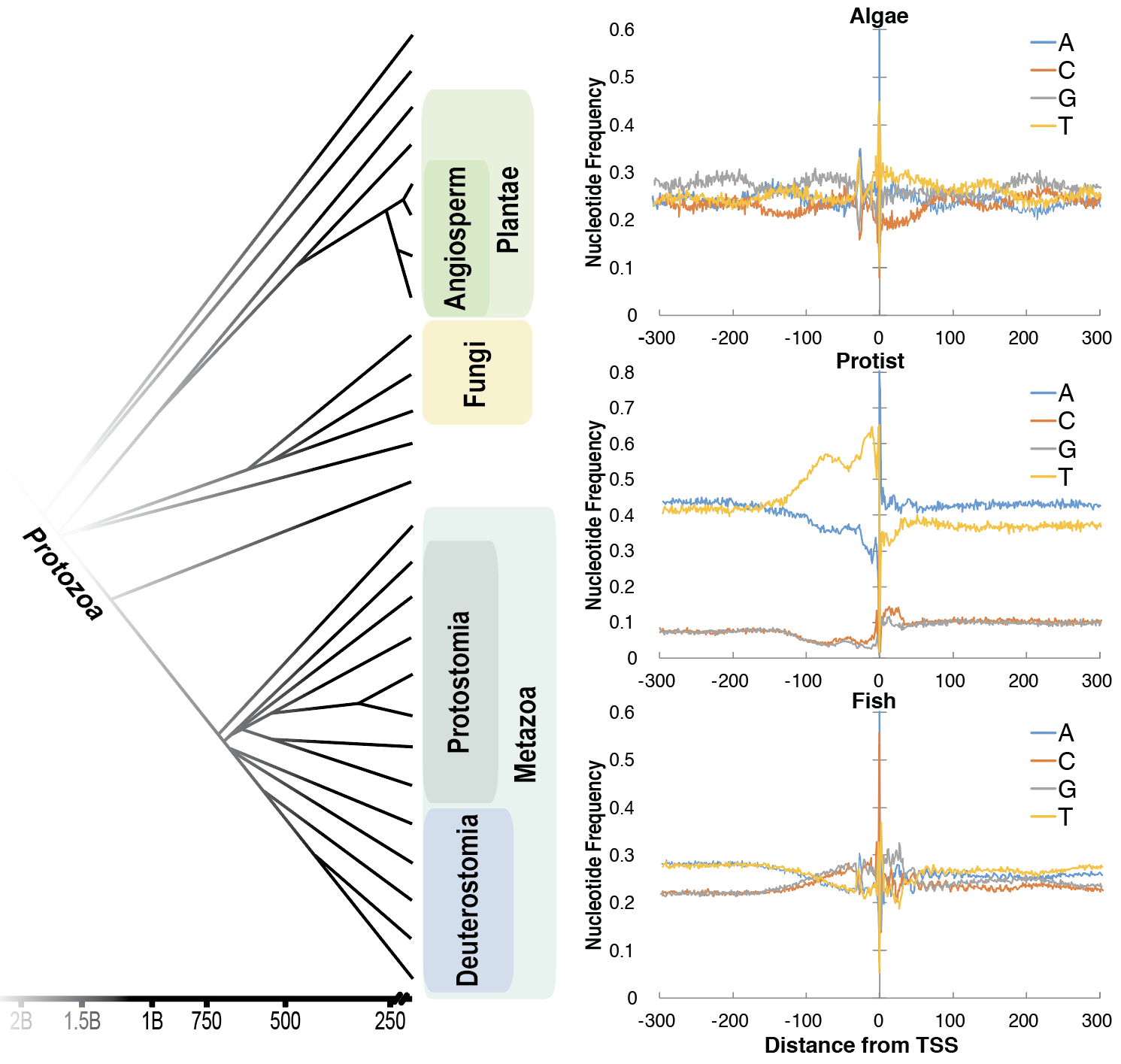
Applying NGS techniques across non-modal organisms to study changes in transcription initiation architecture

We are studying the evolution of eukaryotic transcription by applying advanced NGS techniques across a large number of non-model organisms to study changes in transcription initiation architecture. In close collaboration with Sascha Duttke in the Glass lab at UCSD, we are working to map the precise locations of transcription initiation across a broad range of eukaryotes and understand the diversity and conserved properties of both protein coding and ncRNA promoters across evolution.
Publication
Duttke SH, Guzman C, Chang M, Delos Santos NP, McDonald BR, Xie J, Carlin AF, Heinz S, Benner C. Position-dependent function of human sequence-specific transcription factors Nature. 2024 Jul 17.
Weber AA, Yang X, Mennillo E, Wong S, Le S, Ashley Teo JY, Chang M, Benner CW, Ding J, Jain M, Chen S, Karin M, Tukey RH.Triclosan administration to humanized UDP-glucuronosyltransferase 1 neonatal mice induces UGT1A1 through a dependence on PPARα and ATF4. J Biol Chem. 2024 May 3.
Bali P, Lozano-Pope I, Hernandez J, Estrada MV, Corr M, Turner MA, Bouvet M, Benner C, Obonyo M.TRIF-IFN-I pathway in Helicobacter-induced gastric cancer in an accelerated murine disease model and patient biopsies iScience. 2024 Apr 19.
de Jong TV, Pan Y, Rastas P, Munro D, Tutaj M, Akil H, Benner C, Chen D, Chitre AS, Chow W, Colonna V, Dalgard CL, Demos WM, Doris PA, Garrison E, Geurts AM, Gunturkun HM, Guryev V, Hourlier T, Howe K, Huang J, Kalbfleisch T, Kim P, Li L, Mahaffey S, Martin FJ, Mohammadi P, Ozel AB, Polesskaya O, Pravenec M, Prins P, Sebat J, Smith JR, Solberg Woods LC, Tabakoff B, Tracey A, Uliano-Silva M, Villani F, Wang H, Sharp BM, Telese F, Jiang Z, Saba L, Wang X, Murphy TD, Palmer AA, Kwitek AE, Dwinell MR, Williams RW, Li JZ, Chen H.A revamped rat reference genome improves the discovery of genetic diversity in laboratory rats. Cell Genom. 2024 Apr 10.
Chen H, King FJ, Zhou B, Wang Y, Canedy CJ, Hayashi J, Zhong Y, Chang MW, Pache L, Wong JL, Jia Y, Joslin J, Jiang T, Benner C, Chanda SK, Zhou Y.Drug target prediction through deep learning functional representation of gene signatures Nat Commun. 2024 Feb 29.
Diego JG, Singh G, Jangra S, Handrejk K, Laporte M, Chang LA, El Zahed SS, Pache L, Chang MW, Warang P, Aslam S, Mena I, Webb BT, Benner C, García-Sastre A, Schotsaert M. Breakthrough infections by SARS-CoV-2 variants boost cross-reactive hybrid immune responses in mRNA-vaccinated Golden Syrian hamsters PLoS Pathog. 2024 Jan.
Torre D, Fstkchyan YS, Ho JSY, Cheon Y, Patel RS, Degrace EJ, Mzoughi S, Schwarz M, Mohammed K, Seo JS, Romero-Bueno R, Demircioglu D, Hasson D, Tang W, Mahajani SU, Campisi L, Zheng S, Song WS, Wang YC, Shah H, Francoeur N, Soto J, Salfati Z, Weirauch MT, Warburton P, Beaumont K, Smith ML, Mulder L, Villalta SA, Kessenbrock K, Jang C, Lee D, De Rubeis S, Cobos I, Tam O, Hammell MG, Seldin M, Shi Y, Basu U, Sebastiano V, Byun M, Sebra R, Rosenberg BR, Benner C, Guccione E, Marazzi I. Nuclear RNA catabolism controls endogenous retroviruses, gene expression asymmetry, and dedifferentiation. Mol Cell. 2023 Dec 7.
Doyle MR, Martinez AR, Qiao R, Dirik S, Di Ottavio F, Pascasio G, Martin-Fardon R, Benner C, George O, Telese F, de Guglielmo G.Strain and sex-related behavioral variability of oxycodone dependence in rats. Neuropharmacology. 2023 Oct 1.
Guzman C, Duttke S, Zhu Y, De Arruda Saldanha C, Downes NL, Benner C, Heinz S.Combining TSS-MPRA and sensitive TSS profile dissimilarity scoring to study the sequence determinants of transcription initiation. Nucleic Acids Res. 2023 Aug 25.
Yang X, Weber AA, Mennillo E, Paszek M, Wong S, Le S, Teo JYA, Chang M, Benner CW, Tukey RH, Chen S.Oral arsenic administration to humanizedUDP-glucuronosyltransferase1 neonatal mice induces UGT1A1 through a dependence on Nrf2 and PXR. J Biol Chem. 2023 Mar.
Lam MTY, Duttke SH, Odish MF, Le HD, Hansen EA, Nguyen CT, Trescott S, Kim R, Deota S, Chang MW, Patel A, Hepokoski M, Alotaibi M, Rolfsen M, Perofsky K, Warden AS, Foley J, Ramirez SI, Dan JM, Abbott RK, Crotty S, Crotty Alexander LE, Malhotra A, Panda S, Benner CW, Coufal NG.Dynamic activity in cis-regulatory elements of leukocytes identifies transcription factor activation and stratifies COVID-19 severity in ICU patients. Cell Rep Med. 2023 Feb 21.
Kandel R, Jung J, Syau D, Kuo T, Songster L, Horn C, Chapman C, Aguayo A, Duttke S, Benner C, Neal SE.Yeast derlin Dfm1 employs a chaperone-like function to resolve misfolded membrane protein stress. PLoS Biol. 2023 Jan.
Delos Santos NP, Duttke S, Heinz S, Benner C.MEPP: more transparent motif enrichment by profiling positional correlations NAR Genom Bioinform. 2022 Dec.
Gandhi S, Mitterhoff R, Rapoport R, Farago M, Greenberg A, Hodge L, Eden S, Benner C, Goren A, Simon I.Mitotic H3K9ac is controlled by phase-specific activity of HDAC2, HDAC3, and SIRT1. Life Sci Alliance. 2022 Oct.
Destici E, Zhu F, Tran S, Preissl S, Farah EN, Zhang Y, Hou X, Poirion OB, Lee AY, Grinstein JD, Bloomekatz J, Kim HS, Hu R, Evans SM, Ren B, Benner C, Chi NC. Human-gained heart enhancers are associated with species-specific cardiac attributes Nat Cardiovasc Res. 2022 Sep.
Branche E, Wang YT, Viramontes KM, Valls Cuevas JM, Xie J, Ana-Sosa-Batiz F, Shafee N, Duttke SH, McMillan RE, Clark AE, Nguyen MN, Garretson AF, Crames JJ, Spann NJ, Zhu Z, Rich JN, Spector DH, Benner C, Shresta S, Carlin AF.SREBP2-dependent lipid gene transcription enhances the infection of human dendritic cells by Zika virus. Nat Commun. 2022 Sep 12.
Duttke SH, Montilla-Perez P, Chang MW, Li H, Chen H, Carrette LLG, de Guglielmo G, George O, Palmer AA, Benner C, Telese F.Glucocorticoid Receptor-Regulated Enhancers Play a Central Role in the Gene Regulatory Networks Underlying Drug Addiction. Front Neurosci. 2022 May 16.
Duttke SH, Beyhan S, Singh R, Neal S, Viriyakosol S, Fierer J, Kirkland TN, Stajich JE, Benner C, Carlin AF.Decoding Transcription Regulatory Mechanisms Associated with Coccidioides immitis Phase Transition Using Total RNA. mSystems. 2022 Feb 22.
Martin-Sancho L, Tripathi S, Rodriguez-Frandsen A, Pache L, Sanchez-Aparicio M, McGregor MJ, Haas KM, Swaney DL, Nguyen TT, Mamede JI, Churas C, Pratt D, Rosenthal SB, Riva L, Nguyen C, Beltran-Raygoza N, Soonthornvacharin S, Wang G, Jimenez-Morales D, De Jesus PD, Moulton HM, Stein DA, Chang MW, Benner C, Ideker T, Albrecht RA, Hultquist JF, Krogan NJ, García-Sastre A, Chanda SK. Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy. Nat Microbiol. 2021 Oct.
Shamie I, Duttke SH, Karottki KJC, Han CZ, Hansen AH, Hefzi H, Xiong K, Li S, Roth SJ, Tao J, Lee GM, Glass CK, Kildegaard HF, Benner C, Lewis NE.A Chinese hamster transcription start site atlas that enables targeted editing of CHO cells. NAR Genom Bioinform. 2021 Jul 13.
Lim JY, Duttke SH, Baker TS, Lee J, Gambino KJ, Venturini NJ, Ho JSY, Zheng S, Fstkchyan YS, Pillai V, Fajgenbaum DC, Marazzi I, Benner C, Byun M.DNMT3A haploinsufficiency causes dichotomous DNA methylation defects at enhancers in mature human immune cells J Exp Med. 2021 Jul 5.
Martin-Sancho L, Lewinski MK, Pache L, Stoneham CA, Yin X, Becker ME, Pratt D, Churas C, Rosenthal SB, Liu S, Weston S, De Jesus PD, O'Neill AM, Gounder AP, Nguyen C, Pu Y, Curry HM, Oom AL, Miorin L, Rodriguez-Frandsen A, Zheng F, Wu C, Xiong Y, Urbanowski M, Shaw ML, Chang MW, Benner C, Hope TJ, Frieman MB, García-Sastre A, Ideker T, Hultquist JF, Guatelli J, Chanda SK. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell. 2021 June 17.
Ho JSY, Mok BW, Campisi L, Jordan T, Yildiz S, Parameswaran S, Wayman JA, Gaudreault NN, Meekins DA, Indran SV, Morozov I, Trujillo JD, Fstkchyan YS, Rathnasinghe R, Zhu Z, Zheng S, Zhao N, White K, Ray-Jones H, Malysheva V, Thiecke MJ, Lau SY, Liu H, Zhang AJ, Lee AC, Liu WC, Jangra S, Escalera A, Aydillo T, Melo BS, Guccione E, Sebra R, Shum E, Bakker J, Kaufman DA, Moreira AL, Carossino M, Balasuriya UBR, Byun M, Albrecht RA, Schotsaert M, Garcia-Sastre A, Chanda SK, Miraldi ER, Jeyasekharan AD, TenOever BR, Spivakov M, Weirauch MT, Heinz S, Chen H, Benner C, Richt JA, Marazzi I. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell. 2021 Mar 30.
Flagg MP, Wangeline MA, Holland SR, Duttke SH, Benner C, Neal S, Hampton RY.Inner-nuclear-membrane-associated degradation employs Dfm1-independent retrotranslocation and alleviates misfolded transmembrane-protein toxicity. Mol Biol Cell. 2021 Feb 10.
Cividini F, Scott BT, Suarez J, Casteel DE, Heinz S, Dai A, Diemer T, Suarez JA, Benner CW, Ghassemian M, Dillmann WH.Ncor2/PPARα-Dependent Upregulation of MCUb in the Type 2 Diabetic Heart Impacts Cardiac Metabolic Flexibility and Function. Diabetes. 2021 Mar.
Tu LN, Hsieh L, Kajimoto M, Charette K, Kibiryeva N, Forero A, Hampson S, Marshall JA, O'Brien J, Scatena M, Portman MA, Savan R, Benner C, Aliseda A, Nuri M, Bittel D, Pastuzsko P, Nigam V.Shear stress associated with cardiopulmonary bypass induces expression of inflammatory cytokines and necroptosis in monocytes. JCI Insight. 2021 Jan 11.
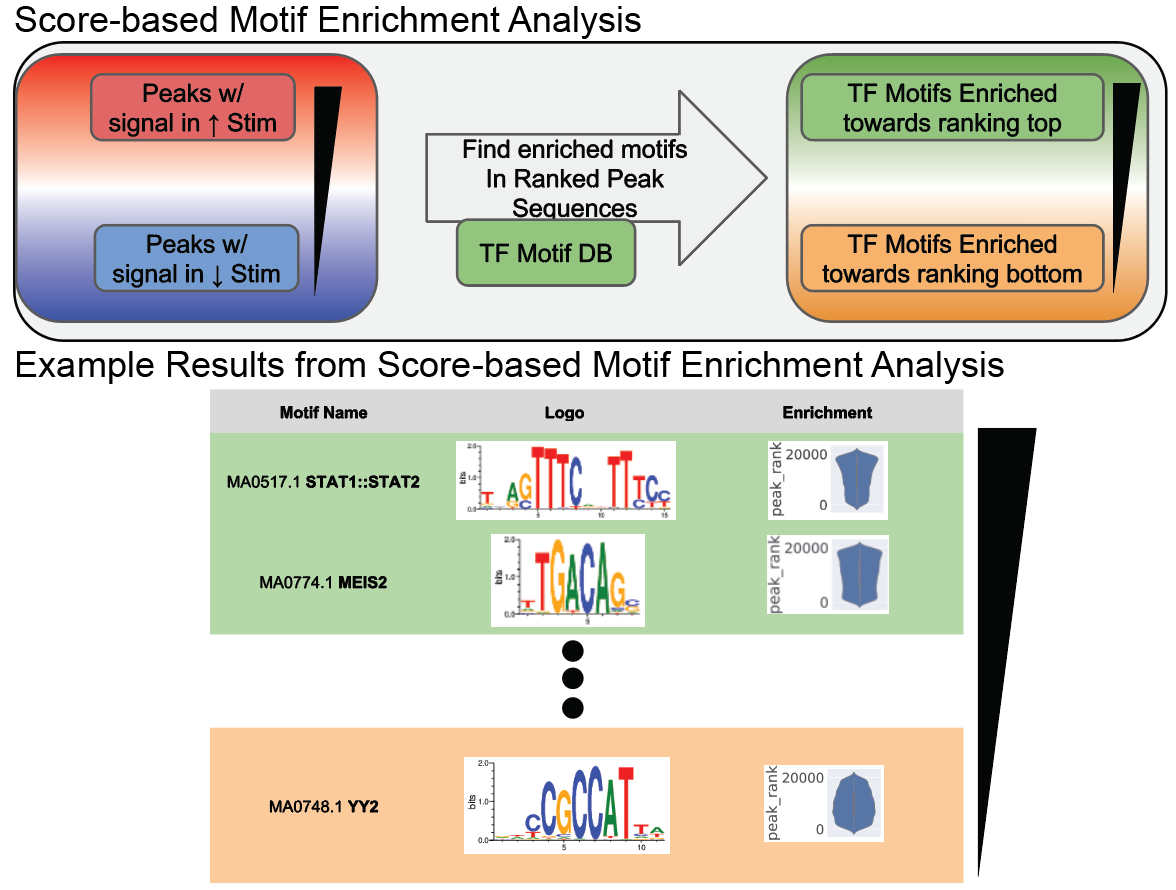
Delos Santos NP, Texari L, Benner C.MEIRLOP: improving score-based motif enrichment by incorporating sequence bias covariates. BMC Bioinformatics. 2020 Sep 16.
Tran V, Ledwith MP, Thamamongood T, Higgins CA, Tripathi S, Chang MW, Benner C, García-Sastre A, Schwemmle M, Boon ACM, Diamond MS, Mehle A. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat Microbiol. 2020 Dec.
Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, Burgstaller-Muehlbacher S, De Jesus PD, Teriete P, Hull MV, Chang MW, Chan JF, Cao J, Poon VK, Herbert KM, Cheng K, Nguyen TH, Rubanov A, Pu Y, Nguyen C, Choi A, Rathnasinghe R, Schotsaert M, Miorin L, Dejosez M, Zwaka TP, Sit KY, Martinez-Sobrido L, Liu WC, White KM, Chapman ME, Lendy EK, Glynne RJ, Albrecht R, Ruppin E, Mesecar AD, Johnson JR, Benner C, Sun R, Schultz PG, Su AI, García-Sastre A, Chatterjee AK, Yuen KY, Chanda SK.Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing Nature. 2020 Oct.
Shen Z, Hoeksema MA, Ouyang Z, Benner C, Glass CK.MAGGIE: leveraging genetic variation to identify DNA sequence motifs mediating transcription factor binding and function. Bioinformatics. 2020 Jul 1.
Ho JSY, Angel M, Ma Y, Sloan E, Wang G, Martinez-Romero C, Alenquer M, Roudko V, Chung L, Zheng S, Chang M, Fstkchyan Y, Clohisey S, Dinan AM, Gibbs J, Gifford R, Shen R, Gu Q, Irigoyen N, Campisi L, Huang C, Zhao N, Jones JD, van Knippenberg I, Zhu Z, Moshkina N, Meyer L, Noel J, Peralta Z, Rezelj V, Kaake R, Rosenberg B, Wang B, Wei J, Paessler S, Wise HM, Johnson J, Vannini A, Amorim MJ, Baillie JK, Miraldi ER, Benner C, Brierley I, Digard P, Łuksza M, Firth AE, Krogan N, Greenbaum BD, MacLeod MK, van Bakel H, Garcìa-Sastre A, Yewdell JW, Hutchinson E, Marazzi I.Hybrid Gene Origination Creates Human-Virus Chimeric Proteins during Infection. Cell. 2020 Jun 25.
Thamamongood T, Aebischer A, Wagner V, Chang MW, Elling R, Benner C, García-Sastre A, Kochs G, Beer M, Schwemmle M. A Genome-Wide CRISPR-Cas9 Screen Reveals the Requirement of Host Cell Sulfation for Schmallenberg Virus Infection. J Virol. 2020 Aug 17.
Roth SJ, Heinz S, Benner C. ARTDeco: automatic readthrough transcription detection BMC Bioinformatics 2020 May 26
Vinckier NK, Patel NA, Geusz RJ, Wang A, Wang J, Matta I, Harrington AR, Wortham M, Wetton N, Wang J, Jhala US, Rosenfeld MG, Benner CW, Shih HP, Sander M. LSD1-mediated enhancer silencing attenuates retinoic acid signalling during pancreatic endocrine cell development Nature Communications 2020 Apr 29
Weiss RJ, Spahn PN, Toledo AG, Chiang AWT, Kellman BP, Li J, Benner C, Glass CK, Gordts PLSM, Lewis NE, Esko JD. ZNF263 is a transcriptional regulator of heparin and heparan sulfate biosynthesis Proc Natl Acad Sci U S A. 2020 Apr 28.
Liu X, Rosenthal SB, Meshgin N, Baglieri J, Musallam SG, Diggle K, Lam K, Wu R, Pan SQ, Chen Y, Dorko K, Presnell S, Benner C, Hosseini M, Tsukamoto H, Brenner D, Kisseleva T. Primary Alcohol‐Activated Human and Mouse Hepatic Stellate Cells Share Similarities in Gene‐Expression Profiles Hepatol Commun. 2020 Feb 6.
Mennillo E, Yang X, Paszek M, Auwerx J, Benner C, Chen S. NCoR1 Protects Mice From Dextran Sodium Sulfate-Induced Colitis by Guarding Colonic Crypt Cells From Luminal Insult Cell Mol Gastroenterol Hepatol. 2020 Feb 7
Liu X, Xu J, Rosenthal S, Zhang LJ, McCubbin R, Meshgin N, Shang L, Koyama Y, Ma HY, Sharma S, Heinz S, Glass CK, Benner C, Brenner DA, Kisseleva T. Identification of Lineage-Specific Transcription Factors That Prevent Activation of Hepatic Stellate Cells and Promote Fibrosis Resolution Gastroenterology 2020 May
Ma HY, Yamamoto G, Xu J, Liu X, Karin D, Kim JY, Alexandrov LB, Koyama Y, Nishio T, Benner C, Heinz S, Rosenthal SB, Liang S, Sun M, Karin G, Zhao P, Brodt P, Mckillop IH, Quehenberger O, Dennis E, Saltiel A, Tsukamoto H, Gao B, Karin M, Brenner DA, Kisseleva T. IL-17 Signaling in Steatotic Hepatocytes and Macrophages Promotes Hepatocellular Carcinoma in Alcohol-Related Liver Disease J Hepatol . 2020 May.
Rodriguez-Frandsen A, Martin-Sancho L, Gounder AP, Chang MW, Liu WC, De Jesus PD, von Recum-Knepper J, Dutra MS, Huffmaster NJ, Chavarria M, Mena I, Riva L, Nguyen CB, Dobariya S, Herbert KM, Benner C, Albrecht RA, García-Sastre A, Chanda SK. Viral Determinants in H5N1 Influenza A Virus Enable Productive Infection of HeLa Cells Journal of Virology 2020 Jan 31.
Duttke SH, Chang MW, Heinz S, Benner C. Identification and dynamic quantification of regulatory elements using total RNA Genome Research. 2019 Nov
Brigidi GS, Hayes MGB, Delos Santos NP, Hartzell AL, Texari L, Lin PA, Bartlett A, Ecker JR, Benner C, Heinz S, Bloodgood BL. Genomic Decoding of Neuronal Depolarization by Stimulus-Specific NPAS4 Heterodimers. Cell. 2019 Oct 3
López-Moyado IF, Tsagaratou A, Yuita H, Seo H, Delatte B, Heinz S, Benner C, Rao A. Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc Natl Acad Sci U S A. 2019 Aug 20
Ramms B, Patel S, Nora C, Pessentheiner AR, Chang MW, Green CR, Golden GJ, Secrest P, Krauss RM, Metallo CM, Benner C, Alexander VJ, Witztum JL, Tsimikas S, Esko JD, Gordts PLSM. ApoC-III ASO promotes tissue LPL activity in the absence of apoE-mediated TRL clearance. J Lipid Res. 2019 Aug
Hishida T, Vazquez-Ferrer E, Hishida-Nozaki Y, Sancho-Martinez I, Takahashi Y, Hatanaka F, Wu J, Ocampo A, Reddy P, Wu MZ, Gerken L, Shaw RJ, Rodriguez Esteban C, Benner C, Nakagawa H, Guillen Garcia P, Nuñez Delicado E, Castells A, Campistol JM, Liu GH, Izpisua Belmonte JC. Mutations in foregut SOX2+ cells induce efficient proliferation via CXCR2 pathway. Protein Cell. 2019 Jul
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019 Apr 3
Wegner M, Diehl V, Bittl V, de Bruyn R, Wiechmann S, Matthess Y, Hebel M, Hayes MG, Schaubeck S, Benner C, Heinz S, Bremm A, Dikic I, Ernst A, Kaulich M. Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife. 2019 Mar 6
Karakus U, Thamamongood T, Ciminski K, Ran W, Günther SC, Pohl MO, Eletto D, Jeney C, Hoffmann D, Reiche S, Schinköthe J, Ulrich R, Wiener J, Hayes MGB, Chang MW, Hunziker A, Yángüez E, Aydillo T, Krammer F, Oderbolz J, Meier M, Oxenius A, Halenius A, Zimmer G, Benner C, Hale BG, García-Sastre A, Beer M, Schwemmle M, Stertz S. MHC class II proteins mediate cross-species entry of bat influenza viruses. Nature. 2019 Mar;567(7746):109-112.
Fonseca GJ, Tao J, Westin EM, Duttke SH, Spann NJ, Strid T, Shen Z, Stender JD, Sakai M, Link VM, Benner C, Glass CK. Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages. Nat Commun. 2019 Jan 24
Carlin AF, Wen J, Vizcarra EA, McCauley M, Chaillon A, Akrami K, Kim C, Ngono AE, Lara-Marquez ML, Smith DM, Glass CK, Schooley RT, Benner C, Shresta S. A longitudinal systems immunologic investigation of acute Zika virus infection in an individual infected while traveling to Caracas, Venezuela. PLoS Negl Trop Dis. 2018 Dec
Carlin AF, Vizcarra EA, Branche E, Viramontes KM, Suarez-Amaran L, Ley K, Heinz S, Benner C, Shresta S, Glass CK. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc Natl Acad Sci U S A. 2018 Sep 25
Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, Marazzi I, García-Sastre A, Shaw ML, Benner C. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018 August 23
Zhu Q, Hoong N, Aslanian A, Hara T, Benner C, Heinz S, Miga KH, Ke E, Verma S, Soroczynski J, Yates JR 3rd, Hunter T, Verma IM. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer Molecular Cell 2018 June 7
Prohaska TA, Que X, Diehl CJ, Hendrikx S, Chang MW, Jepsen K, Glass CK, Benner C, Witztum JL. Massively Parallel Sequencing of Peritoneal and Splenic B Cell Repertoires Highlights Unique Properties of B-1 Cell Antibodies Journal of Immunology 2018 March 1
Neal S, Jaeger PA, Duttke SH, Benner C, K Glass C, Ideker T, Hampton RY. The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins Molecular Cell 2018 Jan 18
Franks TM, McCloskey A, Shokirev MN, Benner C, Rathore A, Hetzer MW. Nup98 recruits the Wdr82–Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells Genes and Development. 2017 Dec 21
Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, Zhong Z, Dhar D, Navas-Molina JA, Xu J, Loomba R, Downes M, Yu RT, Evans RM, Dorrestein PC, Knight R, Benner C, Anstee QM, Karin M. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity Nature. 2017 Nov 16
Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers eLife 2017 Oct 2
Rahnamoun H, Lu H, Duttke SH, Benner C, Glass CK, Lauberth SM. Mutant p53 shapes the enhancer landscape of cancer cells in response to chronic immune signaling Nature Comm. 2017 Sept 29
Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, Tognetti M, Benner CW, Boulton SJ, Saghatelian A, Karlseder J. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN Nature. 2017 Sept 3
Koyama Y, Wang P, Liang S, Iwaisako K, Liu X, Xu J, Zhang M, Sun M, Cong M, Karin D, Taura K, Benner C, Heinz S, Bera T, Brenner DA, Kisseleva T. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest. 2017 Apr 3
Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016 Nov 2.
Hetzel J, Duttke SH, Benner C, Chory J. Nascent RNA sequencing reveals distinct features in plant transcription. Proc Natl Acad Sci USA. 2016 Oct 25.
Nuttle X, Giannuzzi G, Duyzend MH, Schraiber JG, Narvaiza I, Sudmant PH, Penn O, Chiatante G, Malig M, Huddleston J, Benner C, Camponeschi F, Ciofi-Baffoni S, Stessman HA, Marchetto MC, Denman L, Harshman L, Baker C, Raja A, Penewit K, Janke N, Tang WJ, Ventura M, Banci L, Antonacci F, Akey JM, Amemiya CT, Gage FH, Reymond A, Eichler EE. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016 Aug 3.
Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A, Shiau AK, Oegema K. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol. 2016 Jul 18.
Franks TM, Benner C, Narvaiza I, Marchetto MC, Young JM, Malik HS, Gage FH, Hetzer MW. Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev. 2016 May 15.
Rialdi A, Campisi L, Zhao N, Lagda AC, Pietzsch C, Ho JS, Martinez-Gil L, Fenouil R, Chen X, Edwards M, Metreveli G, Jordan S, Peralta Z, Munoz-Fontela C, Bouvier N, Merad M, Jin J, Weirauch M, Heinz S, Benner C, van Bakel H, Basler C, García-Sastre A, Bukreyev A, Marazzi I. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016 May 27.
Ding N, Hah N, Yu RT, Sherman MH, Benner C, Leblanc M, He M, Liddle C, Downes M, Evans RM. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci USA. 2015 Dec 22.
Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Perspectives on Unidirectional versus Divergent Transcription. Mol Cell. 2015 Nov 5.
Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, Moore L, Hunter T, Saghatelian A, Gage FH. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell. 2015 Oct 22.
Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci USA. 2015 Sep 28.
Wu MZ, Chen SF, Nieh S, Benner C, Ger LP, Jan CI, Ma L, Chen CH, Hishida T, Chang HT, Lin YS, Montserrat N, Gascon P, Sancho-Martinez I, Izpisua Belmonte JC. Hypoxia Drives Breast Tumor Malignancy through a TET-TNFα-p38-MAPK Signaling Axis. Cancer Res. 2015 Sep 15.
Puto L, Benner C, Hunter T. The DAXX co-repressor is directly recruited to active regulatory elements genome-wide to regulate autophagy programs in a model of human prostate cancer. Oncoscience Oncoscience. 2015 Apr 17.
Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015 Jun 16.
Wu J, Okamura D, Li M, Suzuki K, Luo C, Ma L, He Y, Li Z, Benner C, Tamura I, Krause MN, Nery JR, Du T, Zhang Z, Hishida T, Takahashi Y, Aizawa E, Kim NY, Lajara J, Guillen P, Campistol JM, Esteban CR, Ross PJ, Saghatelian A, Ren B, Ecker JR, Izpisua Belmonte JC. A spatially defined pluripotent state confers interspecies chimera competency. Nature. 2015 May 21.
Estarás C, Benner C, Jones KA. Wnt-Activin synergy regulates RNAPII pause-release and counteracts a Yap1 elongation block during hESC differentiation. Mol Cell. 2015 Jun 4.
Kashyap MK, Kumar D, Villa R, La Clair JJ, Benner C, Sasik R, Jones H, Ghia EM, Rassenti LZ, Kipps TJ, Burkart MD, Castro JE. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide B. Haematologica. 2015 Apr 10.
Miller MS, Rialdi A, Ho JS, Tilove M, Martinez-Gil L, Moshkina NP, Peralta Z, Noel J, Melegari C, Maestre AM, Mitsopoulos P, Madrenas J, Heinz S, Benner C, Young JA, Feagins AR, Basler CF, Fernandez-Sesma A, Becherel OJ, Lavin MF, van Bakel H, Marazzi I. Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat Immunol. 2015 Mar 30.
Chang AT, Liu Y, Ayyanathan K, Benner C, Jiang Y, Prokop JW, Paz H, Wang D, Fu X, Rauscher FJ, Yang J. An evolutionarily conserved DNA architecture determines target specificity of the Twist-family bHLH transcription factors. Genes Dev. 2015 Mar 15.
Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, Esteban CR, Campistol JM, Yeo GW, Izpisua Belmonte JC. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015 Mar 4.
Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell-type specific enhancers. Nature Reviews. Nat Rev Mol Cell Biol. 2015 Feb 4.
Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Human promoters are intrinsically directional. Mol Cell. 2015 Jan 27.
Hah N, Benner C, Chong LW, Yu RT, Downes M, Evans RM. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc Natl Acad Sci USA. 2015 Jan 6.
Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, Liang G, Tang Z, Mathé E, Benner C, Dubois W, Nelson S, Vian L, Oliveira TY, Jankovic M, Hakim O, Gazumyan A, Pavri R, Awasthi P, Song B, Liu G, Chen L, Zhu S, Feigenbaum L, Staudt L, Murre C, Ruan Y, Robbiani DF, Pan-Hammarström Q, Nussenzweig MC, Casellas R. B Cell Super-Enhancers and Regulatory Clusters Recruit AID Tumorigenic Activity. Cell. 2014 Dec 3.
Kaikkonen MU, Niskanen H, Romanoski CE, Kansanen E, Kivelä AM, Laitalainen J, Heinz S, Benner C, Glass CK, Ylä-Herttuala S. Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization. Nucleic Acids Res. 2014 Nov 10.
Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. Function of a Foxp3 cis-Element in Protecting Regulatory T Cell Identity. Cell. 2014 Aug 14.
Benner C, van der Meulen T, Cacéres E, Tigyi K, Donaldson CJ, Huising MO. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014 Jul 22.
Liu GH, Suzuki K, Li M, Qu J, Montserrat N, Tarantino C, Gu Y, Yi F, Xu X, Zhang W, Ruiz S, Plongthongkum N, Zhang K, Masuda S, Nivet E, Tsunekawa Y, Soligalla RD, Goebl A, Aizawa E, Kim NY, Kim J, Dubova I, Li Y, Ren R, Benner C, del Sol A, Bueren J, Trujillo JP, Surralles J, Cappelli E, Dufour C, Esteban CR, Izpisua Belmonte JC. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat Commun. 2014 Jul 7.
Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao HK, Benner C, Esteban CR, Jin Y, Liu GH, Li Y, Izpisua Belmonte JC. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014 Jul 3.
Crotti A, Benner C, Kerman BE, Gosselin D, Lagier-Tourenne C, Zuccato C, Cattaneo E, Gage FH, Cleveland DW, Glass CK. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci. 2014 Apr.
Benner C, Konovalov S, Mackintosh C, Hutt KR, Stunnenberg R, Garcia-Bassets I. Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions. PLoS Genet. 2013 Nov.
Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013 Nov 28.
Almenar-Queralt A, Kim SN, Benner C, Herrera CM, Kang DE, Garcia-Bassets I, Goldstein LS. Presenilins Regulate Neurotrypsin Gene Expression and Neurotrypsin-dependent Agrin Cleavage via Cyclic AMP Response Element-binding Protein (CREB) Modulation. J Biol Chem. 2013 Dec 6.
Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013 Nov 28.
Gu Y, Liu GH, Plongthongkum N, Benner C, Yi F, Qu J, Suzuki K, Yang J, Zhang W, Li M, Montserrat N, Crespo I, Del Sol A, Esteban CR, Zhang K, Belmonte JC. Global DNA methylation and transcriptional analyses of human ESC-derived cardiomyocytes. Protein Cell. 2013 Aug 27.
Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013 Aug 8.
Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013 Jun 27.
Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Längst G, Benner C, Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013 Jul.
Mansson R, Welinder E, Åhsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci USA. 2012 Dec 18.
Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in nuclear positioning of genes and intra- and inter-domain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012 Oct 14.
Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci USA. 2012 Jun 12.
Pham TH, Benner C, Lichtinger M, Schwarzfischer L, Hu Y, Andreesen R, Chen W, Rehli M. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012 Jun 14.
Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, Cheng CS, Luna R, Ludka C, Sasik R, Garcia-Bassets I, Hoffmann A, Subramaniam S, Hardiman G, Rosenfeld MG, Glass CK. Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes. PLoS Genet. 2011 Dec.
Mercer EM, Lin YC, Benner C, Jhunjhunwala S, Dutkowski J, Flores M, Sigvardsson M, Ideker T, Glass CK, Murre C. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011 Sep 23.
Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011 May 15.
Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK. SRF utilizes distinct promoter and enhancer-based mechanisms to regulate the macrophage-specific program of cytoskeletal gene expression. Mol Cell Biol. 2010 Dec 6.
Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010 Nov 24.
Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010 Aug 30.
Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010 Jul 22.
Wiesner P, Choi SH, Almazan F, Benner C, Huang W, Diehl CJ, Gonen A, Butler S, Witztum JL, Glass CK, Miller YI. Low doses of lipopolysaccharide and minimally oxidized low-density lipoprotein cooperatively activate macrophages via nuclear factor kappa B and activator protein-1: possible mechanism for acceleration of atherosclerosis by subclinical endotoxemia. Circ Res. 2010 Jul 9.
Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010 Jul.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010 May 28.
Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, Rehli M. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer. Cancer Res. 2010 Feb 15.
El Chartouni C, Benner C, Eigner M, Lichtinger M, Rehli M. Transcriptional effects of Colony-stimulating factor-1 in mouse macrophages. Immunobiology. 2009 Sep 14.
Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009 Mar 15.
Young JA, Johnson JR, Benner C, Yan SF, Chen K, Le Roch KG, Zhou Y, Winzeler EA. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics. 2008 Feb 7.
Maurya MR, Benner C, Pradervand S, Glass C, Subramaniam S. Systems biology of macrophages. Adv Exp Med Biol. 2007; 598:62-79.
Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, Watt MJ, Benner C, Febbraio MA, Nguyen AK, Folian B, Subramaniam S, Gonzalez FJ, Glass CK, Ricote M. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007 June.
Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA. 2007 Mar 20.
Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006 May.
Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005 Sep 9.
Barrera L, Benner C, Tao YC, Winzeler E, Zhou Y. Leveraging two-way probe-level block design for identifying differential gene expression with high-density oligonucleotide arrays. BMC Bioinformatics. 2004 Apr 20.
HOMER is a suite of NGS and genomics analysis tools for the processing and integrated analysis of a large number of genomics assays. Although initially designed to analyze ChIP-seq data, it has been expanded over the better part of a decade to include support for a number of emerging NGS technologies, including GRO-seq and Hi-C. HOMER also includes a powerful de novo motif discovery algorithm and associated routines to integrate regulatory motif information with quantitative NGS results.
Features:
Reference:
(Original) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities Mol Cell. 2010 May 28.
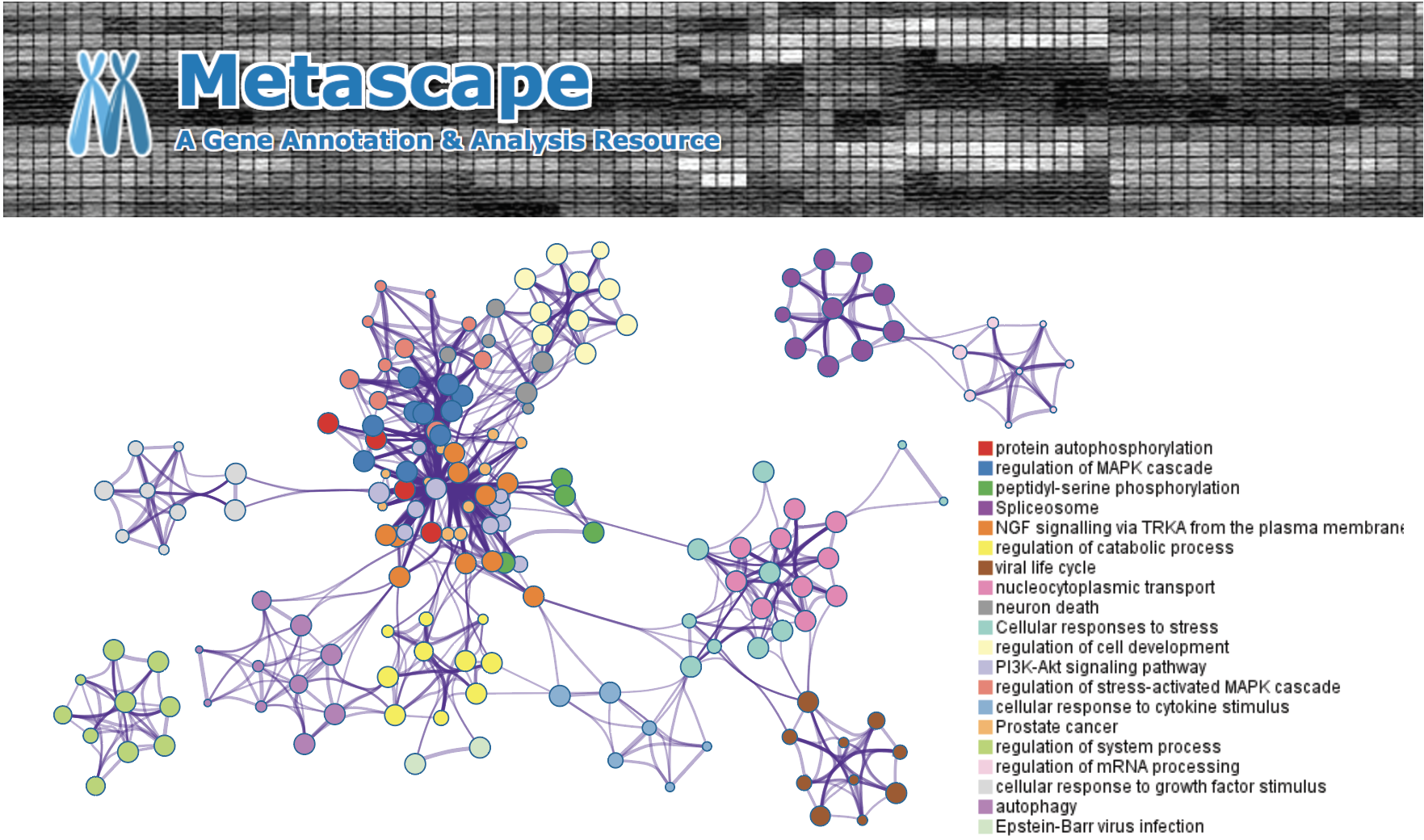
Metascape is a web application composed of a set of reliable, productive and intuitive tools that help researchers analyze gene/protein lists for biological function. In addition to performing Gene Ontology and pathway enrichment, Metascape looks for complexes in protein-protein interaction networks, and is capable of performing a number of annotation and identifier conversion tasks. Metascape is also capable of analyzing several gene lists at the same time to identify similarities and differences in gene lists. Metascape development is led by Yingyao Zhou at the Genomics Institute of the Novartis Research Foundation in collaboration with the Benner lab and the Chanda lab at the Sanford Burnham Prebys Medical Discovery Institute.
Features:
Reference: Metascape provides a biologist-oriented resource for the analysis of systems-level datasets Nat Commun. 2019 Apr 3.
ARTDeco (Automatic Readthrough Transcription DEteCtiOn) ARTDeco is a tool to detect and analyze multiple features of readthrough transcription from RNA-seq and other next-generation sequencing (NGS) assays that profile transcriptional activity. Several diseases (e.g. influenza) and biological processes (e.g. stress response) target transcription termination to misregulate gene expression. Disruption of transcription termination leads to readthrough transcription past the 3′ end of genes, which can result in novel transcripts, changes in epigenetic states and altered 3D genome structure. ARTDeco robustly quantifies the global severity of readthrough phenotypes, and reliably identifies individual genes that fail to terminate (readthrough genes), are aberrantly transcribed due to upstream termination failure (read-in genes), and novel transcripts created as a result of readthrough (downstream of gene or DoG transcripts).
Features:
Reference: ARTDeco: automatic readthrough transcription detection BMC Bioinformatics. 2020 May 26
MEIRLOP MEIRLOP is a new approach to motif analysis that determines enrichment of TF binding motifs in a list of scored regulatory regions. It uses logistic regression while simultaneously attempting to correct for biases in sequence content (i.e. CpG Islands) to accurately identify motifs that strongly associate with scores assigned to sequences/regulatory elements.
Features:
Reference: MEIRLOP: improving score-based motif enrichment by incorporating sequence bias covariates BMC Bioinformatics. 2020 Sep 16
MEPP MEPP is a tool that can be used to investigate the position-dependent association of a motif with a given phenotypic trait, providing both quantitative analyses and visualizations. Given a scored set of sequences (similar to MEIRLOP) with a fixed anchor point, MEPP will correlate motif presense with sequence score as a function of distance along the sequences of interest. This is useful for determining if a TF motif has a functional impact at specific distance relative to an important feature (e.g. the TSS, or a splice site, etc.)
Features:
Reference: MEPP: more transparent motif enrichment by profiling positional correlations. NAR Genom Bioinform. 2022 Oct 17.

Capped short (cs)RNA-seq is a method to map “initiating transcripts” in eukaryotic cells, providing quantitative, single-nucleotide resolution maps of transcription initiation throughout the genome. The general idea is that it isolates 20-60nt RNAs that have a 5'cap, which are mostly found associated with active RNAPII at regulatory elements. The technique builds on other short RNA sequencing approaches, and produces data that is similar to true nascent RNA TSS mapping methods like GRO-cap, PRO-cap, and 5’GRO-seq. csRNA-seq is a [relatively] simple assay, and starts from total RNA, making it easy to apply to a variety of different types of samples.
Features:
Reference: Identification and dynamic quantification of regulatory elements using total RNA Genome Res. 2019 Nov

The ability to profile TSS using a MPRA provides a powerful platform to investigate the impact that regulatory sequence has on the regulation of transcription. Not only does it provide information about the strength/frequency of transcription initiation, it provides positional information for which nucleotides in the reporter drive initiation. Carlos Guzman (Heinz Lab) and Sascha Duttke extended previous designs and worked out the protocols to reliably perform TSS mapping using a massively parallel reporter assay that uses a barcode placed 3’ to the regulatory sequence being interrogated. The assay can work well using libraries with up to 18,000 unique regulatory sequences, which can be synthetically designed and synthesized as an oligo pool, enabling researchers to assay nearly any sequence imaginable.
Reference: Combining TSS-MPRA and sensitive TSS profile dissimilarity scoring to study the sequence determinants of transcription initiation Nucleic Acids Res. 2023 Aug 25.


Dr. Benner is a bioinformatics and genomics scientist fascinated by how our cells decode the regulatory DNA in our genome to orchestrate transcriptional responses. The laboratory develops computational and experimental technologies to study how transcription is regulated, analyzing how regulatory DNA, transcription, epigenetics, and 3D genome structure exert influence on one another. The laboratory leverages experimental methods to profile transcription initiation in the genome (e.g. csRNA-seq, TSS-MPRA), which provide sensitive measurements of regulatory element activity at single-nucleotide resolution. The unique properties of these approaches are then used to model how transcriptional networks respond to inflammatory stimulation or other pathological conditions and monitor the impact that non-coding genetic variation plays in perturbing transcriptional responses. Dr. Benner’s interests also include studying the evolution of eukaryotic transcription by applying advanced NGS techniques across many non-model organisms to reveal broad principles guiding transcriptional processes.
Dr. Benner received his BS in Bioengineering at UCSD before joining the laboratory of Dr. Christopher Glass to pursue his Ph.D. in Bioinformatics. As a graduate student, Chris developed the DNA motif discovery algorithm behind HOMER and began developing next-generation sequencing (NGS) analysis tools to study transcription factors and epigenetic modifications across the genome using ChIP-seq data. As a post-doc Chris continued to develop integrative analysis tools to analyze nascent RNA (GRO-seq) and genome structure (Hi-C) to study enhancer transcription and hematopoietic development and innate immunity. After his post-doc, Chris served for three and a half years as the Director of Integrative Genomics and Bioinformatics Core at the Salk Institute, where he worked with dozens of laboratories to advance their research using NGS technologies, gaining experience analyzing different types of genomics data in fields spanning cancer, cellular development, and evolution. Chris was appointed as an Assistant Professor at UCSD in 2016. The current laboratory continues in this spirit, collaborating with multiple groups to study viral host-pathogen interactions, cellular development, chronic inflammatory diseases, and substance abuse. They also continue to develop genomics software, exemplified by HOMER and the pathway enrichment tool Metascape, that can be leveraged by the scientific community to improve the interpretation of genomics data.















cbenner@ucsd.edu
(858) 534-9449
9500 Gilman Drive MC 0640
La Jolla, CA 92093-0640
George Palade Laboratories (CMM west)
GPL Room 105H
School of Medicine
University of California, San Diego