
HOMER
Software for motif discovery and next-gen sequencing analysis
RNA Motif Analysis
HOMER was not originally designed with RNA in mind, but it
can be used
to successfully analyze data for RNA motifs. By RNA
motifs, we
mean short sequence elements in RNA sequences akin to DNA
motifs, not
structural elements such as hairpins and stuff like
that. For
example, HOMER can be used to successfully determine miRNA
seeds in
sets of co-regulated mRNAs, or RNA binding elements in
CLIP-Seq data.The "-rna" option can be used with findMotifs.pl and findMotifsGenome.pl, resulting in + strand only motif searching and motif display/matching with "U" instead of "T". HOMER does not contain a list of well known "RNA motifs" yet, so no "known motif" analysis is performed. If using FASTA files, please use "T" (normal DNA encoding) in the input files for now.
Analyzing Co-regulated Gene Lists for RNA motifs
HOMER contains preconfigured
PROMOTER sets comprised of RefSeq mRNA sequences, or only
the 5' and 3'
UTRs. These are useful for analyzing gene lists for
motifs in
their mRNAs. To run the analysis, us findMotifs.pl
with a mRNA
PROMOTER set, and options for RNA motifs will be
automatically set.
You don't actually need to specify -rna for this case since with the use of "human-mRNA" it's understood. Anyway, the output will look something like this:
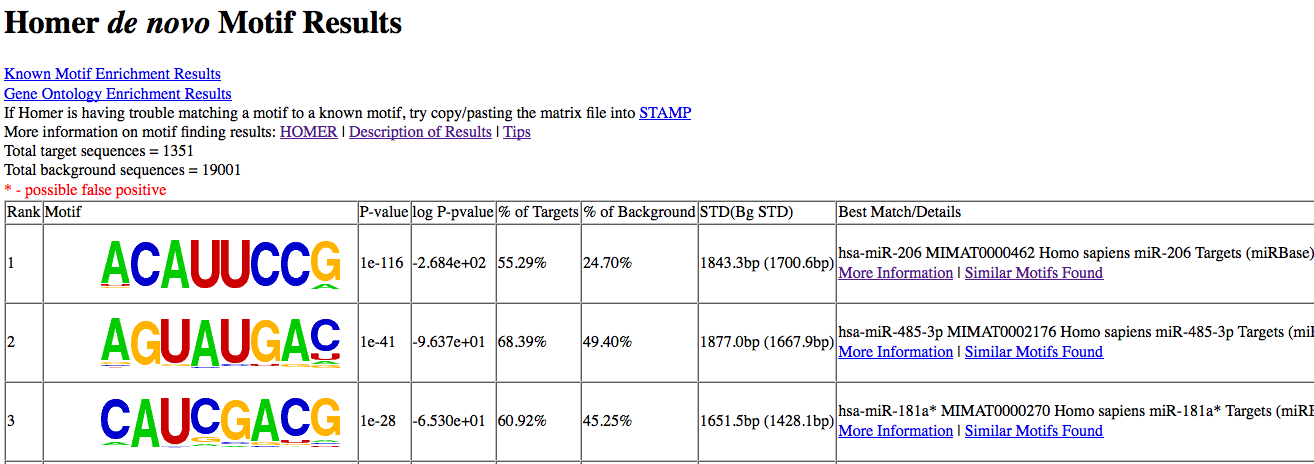
For now, HOMER will try to match the results to the human list of miRNA seeds (from miRBase):

In this case, the motif matches the miR-1 consensus seed (which is shared by miR-206 and miR-613).
There are two RNA specific options for findMotifs.pl in rna mode:
findMotifs.pl
mir1-downregulated.genes.txt
human-mRNA
MotifOutput/ -rna -len 8
You don't actually need to specify -rna for this case since with the use of "human-mRNA" it's understood. Anyway, the output will look something like this:

For now, HOMER will try to match the results to the human list of miRNA seeds (from miRBase):

In this case, the motif matches the miR-1 consensus seed (which is shared by miR-206 and miR-613).
There are two RNA specific options for findMotifs.pl in rna mode:
-min
<#> : minimum length of mRNA to consider
(basically
removes extremely short mRNA sequences from the
analysis)
-max <#> : maximum length of mRNA to consider (removes really long RNAs from the analysis)
-max <#> : maximum length of mRNA to consider (removes really long RNAs from the analysis)
Analyzing CLIP-Seq for RNA motifs
HOMER can analyze
strand-specific
genomic regions for motifs, such as the regions that would
be defined
from CLIP-Seq. To do this, just run
findMotifsGenome.pl using the
"-rna" flag (make
sure your
regions are strand specific!!). For now, HOMER just
uses the same
random genomic background used for ChIP-Seq motif
finding. You
could imagine that a better RNA motif finding background
would be RNA,
i.e. strand specific exon/intron sequences. You'd be
right, but
managing this with respect to the different experiments
(i.e. intronic
binding vs. mRNA binding vs. non-coding RNA binding) is
tricky and for
now left up to the user (you can specify your own strand
specific
background with "-bg
<peak/BED
file>"). Trying this with FOX CLIP-Seq
data:
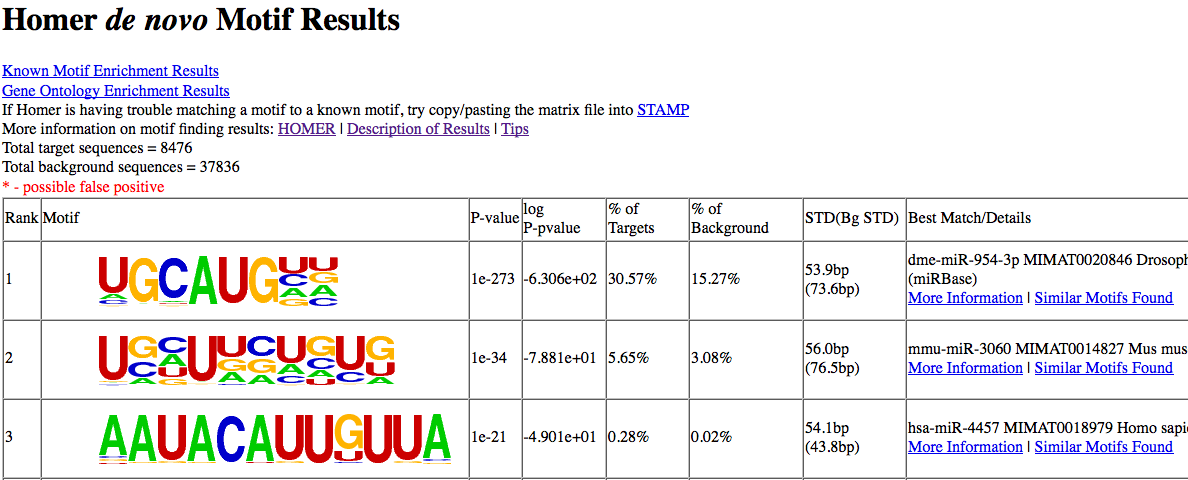
This will give the following results (which resembles a UGCAUG FOX motif):
findMotifsGenome.pl
fox2.clip.bed
hg17
MotifOutput -rna
This will give the following results (which resembles a UGCAUG FOX motif):


Can't figure something out? Questions, comments, concerns, or other feedback:
cbenner@ucsd.edu