
HOMER
Software for motif discovery and ChIP-Seq analysis
Practical Tips to Motif Finding with HOMER
Below are some general tips for getting the most out of you motif analysis when using HOMER. Be sure to look over this section about judging motif quality!What to do if motif finding takes too long...
Ctrl+C... If you are using
reasonable parameters (see next section), it shouldn't take more than
an hour or so, and in most cases much less.
Choosing the length of motifs to find
It's almost always a good idea to
start with the default parameters. Resist the urge to find motifs
larger than 12 bp the first time around. Longer motifs will show
up as different short motifs when finding shorter motifs. If
there aren't any truly significant motifs when looking at short motifs,
it is unlikely that you will find good long motifs either. And it
doesn't take much time to check for short motifs.
i.e. -S 50 -len 8,10
Once you do find motifs that look promising, try looking for longer motifs ("-len 12,14").
If searching for motifs >12 or 14 bp (i.e. 20 bp motifs), try to do the following:
- try to reduce the number of target sequences to include only high quality sequences (such as "focused" peaks or peak with the highest peak scores).
- try limiting the length of sequences used (i.e. "-size 50" when using findMotifsGenome.pl)
- try limiting the total number of background sequences (i.e. "-N 20000" when using findMotifsGenome.pl)
How many sequences can HOMER handle?
In theory, a lot (i.e.
millions). It has been designed to work well with ~10k target
sequences and 50k background sequences. If you are using a large
number of sequences with findMotifs.pl,
you
many
want
to
use
the "-b"
option, which switches to the cumulative
binomial distribution for motif scoring, which is faster to
calculate and gives essentially the same results when using large
numbers of sequences. The binomial is used by default in findMotifsGenome.pl. (I guess it
should be called BOMER !?).
Choosing background sequences
Most of the methods in HOMER
attempt to select the proper background for you, but in some cases this
doesn't work. Normally, HOMER attempts to normalize the CpG
content in target and background sequences. If you believe
normalizing the overall G+C content (GC) is better, use the option "-gc" when performing motif finding
with either findMotifs.pl or findMotifsGenome.pl.
In some cases the user may have a better idea of what the background should be, so HOMER offers the following options:
In some cases the user may have a better idea of what the background should be, so HOMER offers the following options:
Promoters: When using
analyzing promoters with findMotifs.pl,
if
you
wish
to
use
a specific set of promoters as background, place
them in a text file (1st column is the ID) and use the "-bg <background IDs file>"
option. Genes found in the target and background will be removed
from the background set so that they don't cancel out each other.
Examples:
- Use expressed genes from a microarray as background
- Use only genes represented on the microarray as background
- Specify peaks common to two cell types as background when trying to find motifs specific to a set of cell-type specific peaks - this will help cancel out the primary motif and reveal the co-enriched motifs
- If peaks are near Exons, specify regions on Exons as
background to remove triplet bias.
You man also want to disable
CpG/GC normalization depending on how you selected your background,
which can be done with "-noweight".
How to
Judge the Quality of the Motifs Found
WARNING: Because this is the hardest
thing for people to understand, I'll say it again here. HOMER
will print the best guess for the motif next to the motif results, but
before you tell your adviser that your factor is enriched for that
motif, it is highly recommended that you look at the alignment!!!
Here
is
an
example
of what might be going on:

In this case, HOMER has identified YY1 as the "best guess" match for this de novo motif. Well, lets click on "More Information" and see what's up:
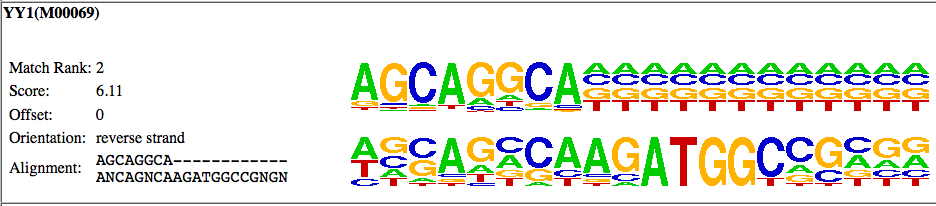
As you can see in this case, the motif aligns to the edge of the known YY1 motif, and not to the core of the YY1 motif (CAAGATGGC). This doesn't mean that the YY1 motif is not enriched in your data, but unless there are other motif results that show enrichment of the other parts of the YY1 motif, it is not likely that the YY1 motif is enriched in your data set.
And as always, remember that HOMER is a de novo motif tool!!! Even though HOMER will guess the best match, if it is a novel motif, your don't want to trust that match anyway. Hence, the you can see the importance of viewing the alignment and getting a feel for what evidence exists either for or against this assignment.
There are many cases where HOMER will find motifs with very low p-values, but the motifs might look "suspicious". Poor quality motifs can be loosely classified into the following groups:

In this case, HOMER has identified YY1 as the "best guess" match for this de novo motif. Well, lets click on "More Information" and see what's up:

As you can see in this case, the motif aligns to the edge of the known YY1 motif, and not to the core of the YY1 motif (CAAGATGGC). This doesn't mean that the YY1 motif is not enriched in your data, but unless there are other motif results that show enrichment of the other parts of the YY1 motif, it is not likely that the YY1 motif is enriched in your data set.
And as always, remember that HOMER is a de novo motif tool!!! Even though HOMER will guess the best match, if it is a novel motif, your don't want to trust that match anyway. Hence, the you can see the importance of viewing the alignment and getting a feel for what evidence exists either for or against this assignment.
There are many cases where HOMER will find motifs with very low p-values, but the motifs might look "suspicious". Poor quality motifs can be loosely classified into the following groups:
Low Complexity Motifs:
These types of motifs tend to
show preference for same collection of 1, 2, 3, or 4 nucleotides in
each position and are typically very degenerate. For example:
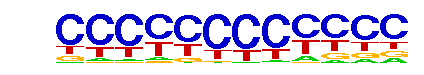
These motifs typically arise when a systematic bias exists between
target and background sequence sets. Commonly they will be very
high in GC-content, in which case you may want to try adding "-gc" to your motif finding command
to normalize by total GC-content instead of CpG-content.
Other times this will come up when analyzing sequences for various genomic features that have not been controlled for in the background - for example, comparing sequences from promoters to random genomic background sequences in some organisms will show preferences for purines or pyrimidines. HOMER is very sensitive, so if there is a bias in the composition of the sequences, HOMER will likely pick it up.

Other times this will come up when analyzing sequences for various genomic features that have not been controlled for in the background - for example, comparing sequences from promoters to random genomic background sequences in some organisms will show preferences for purines or pyrimidines. HOMER is very sensitive, so if there is a bias in the composition of the sequences, HOMER will likely pick it up.
Simple Repeat Motifs:
Some times motifs will show
repeats of certain patterns:

Usually motifs like this will be accompanied by several other motifs
looking highly similar. Unless there is a good reason to believe
these may be real, it's best to assume there is likely a problem with
the background. These can arise if your target sequences are
highly enriched on exons (think triplets) and other types of sequences,
and if "-gc" doesn't help, you
may have to think hard about the types of sequences that you are trying
to analyze and try to match them. (i.e. Promoters vs. Promoters,
Exons vs. Exons etc.)

Small Quantity Motifs / Repeats:
These are a little harder to
explain. These look like real motifs but are found in an
incredibly low percentage of targets - i.e. like an oligo or part of a
repeat that is in a couple of the target sequences that appears as a
significant motif. Statistically speaking they are enriched, but
likely not real. These are the biggest problem when looking for
motifs in promoters from a small list of regulated genes. In
principle, in a motif is present in less
than
5%
of
the
targets sequences, there may be a problem.
Leftover Junk:
These are motifs that appear in
your lower in your results list after you've discovered high quality
motifs. If an element is highly enriched in your sequences, HOMER
will (hopefully) find it, mask it, and then continue to look for
motifs. In this case, many of the other motifs that HOMER finds
will be offsets or degenerate versions of highly enriched motif(s)
found
at the beginning. For example (another PU.1 example):
The top motif identified:
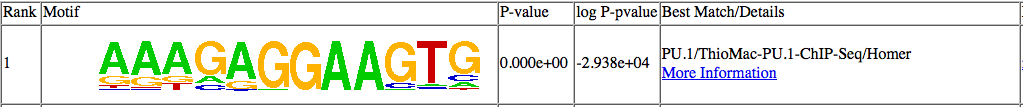
Examples further down the list:



This are not necessarily negative results, but they should be place in
context. This commonly happens in ChIP-Seq data sets where the
immunoprecipitated protein is highly expressed and binds strongly a ton
of binding sites. These "other" motifs are likely also capable of
binding PU.1 and probably represent low affinity binding sites, but
giving them too much individual attention is not recommended in this
context given they are motifs that have been constructed using leftover
oligos in the motif finding process that didn't make it into the most
highly enrichment motifs. A safer way to approach these elements
is to repeat the motif finding procedure with regions lacking the top
motif, or by adding "-mask <motif
file>" to the motif finding command to cleanly mask the top
motif from the motif finding procedure.
Examples further down the list:




Can't figure something out? Questions, comments, concerns, or other feedback:
cbenner@ucsd.edu