HOMER
Software for motif discovery and next-gen sequencing analysis
Hi-C Analysis Tips
Hi-C is a beast of an assay. There is a lot of
information in each run, and the size and complexity of the
experiments can stretch the limits of your computer
too. Below are some tips and "best practices" to help
get Hi-C analysis with HOMER to turn out successful.Resolution and Contact Matrices
Keep in mind that the size of the region you are looking divided by the resolution will yield the total size of your matrix. For example, mouse chr1 is about 200 Mb long. If you make matrix with chr1 at 100kb resolution, you matrix will be 2000x2000 in size. Even if one pixel is used to visualize each data point in the matrix, it's not going to fit on your computer screen. It will also be a very large file (tens of megabytes). If you try to visualize chr1 at 10kb resolution, your matrix will be 20000x20000, which in all likely hood will eat up all of your memory and disk. Homer has a checkpoint to warn you about this, but in general, if you want to visualize something at high resolution, use a smaller region.
Resolution and Running Time
The smaller the size of the regions used for analysis, the longer everything is going to take. In general, the analysis time scales O(n2) with the number of regions analyzed, so analysis with a resolution of 10000 will take approximately 4x longer than analysis with a resolution of 20000.
In practice it's much better to start with a low resolution (i.e. 100kb), and if everything works out, try a higher resolution (i.e. 10kb).
How small can the Resolution/Super Resolution Go?
Is it possible to analyze my data at 5kb resolution? What about 1kb or 250 bp resolution?
Number of CPUs and Memory
Several of the HOMER Hi-C programs support multiple processors to help speed up the computation. Unfortunately, as of now this works at the chromosome level. This means that if you use "-cpu 3" with the analyzeHiC command, it will spawn 3 threads to handle chr1, chr2, and chr3 at the same time. As each chromosome finishes, it will start on chr4 next, and so on. Ideally the parallel code would be deeper in the analysis (i.e. analyze chr1 at 3x speed), but I haven't had time to go back in and re-butcher everything.
On important practical consideration: The total data for each chromosome is read into memory! If your experiment is very large, this may cause problems. For example, lets say you have 16 CPUs on you computer, so you run analyzeHiC with "-cpu 16". The data for each of the first 16 chromosomes will then be read into memory... If the experiment is very large (say a billion reads), you may run out of memory, particularly if you only have 50-100Gb). In these cases, you may be forced to use less CPUs...
Impact of Genomic Rearrangements and Copy Number Variants on Analysis
Genomic rearrangements in
particular can wreak havoc on Hi-C analysis.
Ideally, Hi-C reads should be mapped to the actual genome
of the cells being analyzed. However, is option is
not always available and difficult to perform in
practice. How do you know if your cells have any
major genome altercations? I'd recommend creating a
genome-wide interaction matrix at 1Mb resolution.
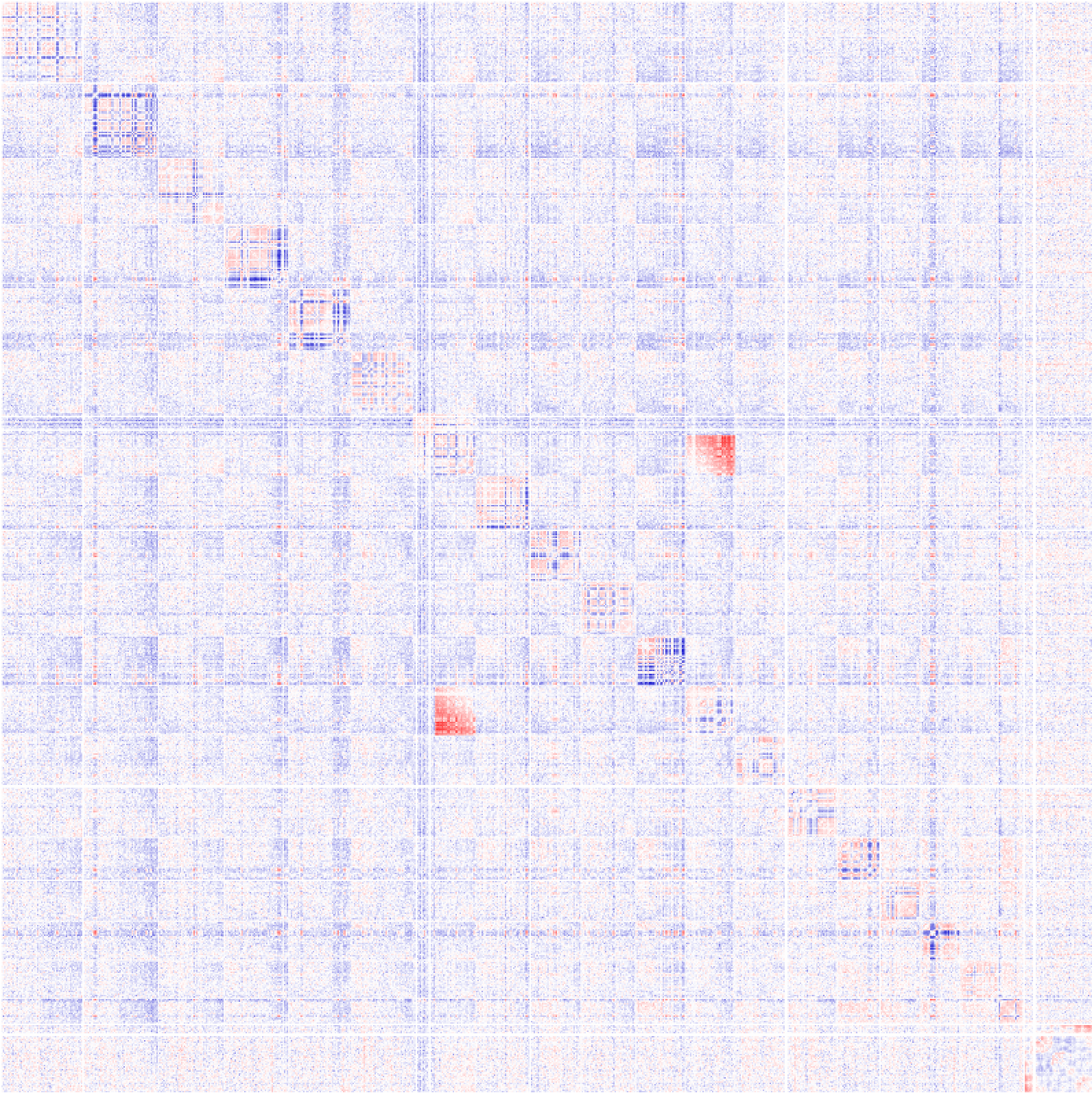
For exampe, consider the following:
How does this effect your analysis? Ideally, the coordinates of the reads could be adjusted to reflect the real genome present in the cell. This is tricky (nearly impossible due to the potential heterozygosity of the rearrangements). The best (or easiest) course of action is to disregard significant interactions and results from these regions (i.e. remove inter-chromosomal interactions between chr7 and chr12 in this case). I will try to update this sections with more tricks and techniques as we encounter more data of this nature.
analyzeHiC HiCexp1/ -res 1000000 -cpu 8 > outputMatrix.txtThe resulting matrix might look something like this:
Here you can see a large block of signal between chr7 and chr12. From this, its a good bet that the latter portion of chr7 is ligated to the end of chr12. You can't necessarily "prove" these cell have a translocation from Hi-C data, but it's likely that this is the case.
How does this effect your analysis? Ideally, the coordinates of the reads could be adjusted to reflect the real genome present in the cell. This is tricky (nearly impossible due to the potential heterozygosity of the rearrangements). The best (or easiest) course of action is to disregard significant interactions and results from these regions (i.e. remove inter-chromosomal interactions between chr7 and chr12 in this case). I will try to update this sections with more tricks and techniques as we encounter more data of this nature.

Can't figure something out? Questions, comments, concerns, or other feedback:
cbenner@ucsd.edu